SEdb 2.0
A comprehensive Super-Enhancer database of human and mouse
What is SEdb 2.0?
Super-enhancers are a large cluster of transcriptionally active enhancers enriched in enhancer-associated chromatin characteristics. Compared to typical enhancers, super-enhancers are larger, exhibit higher transcription factor density, and are frequently associated with key lineage-specific genes that control cell state and differentiation in somatic cells.
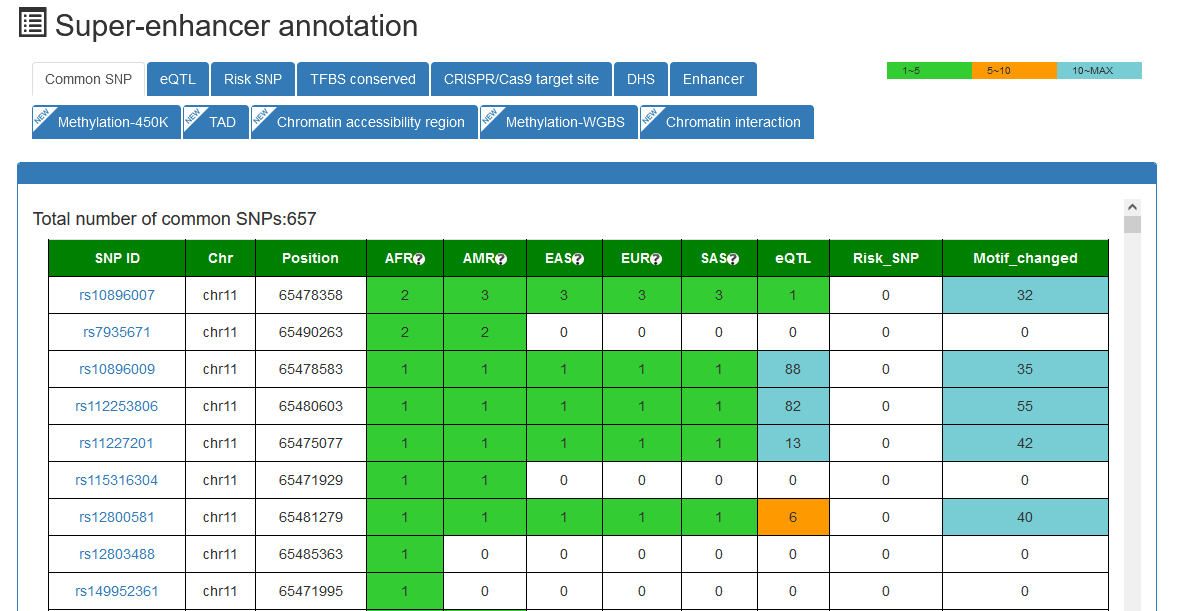
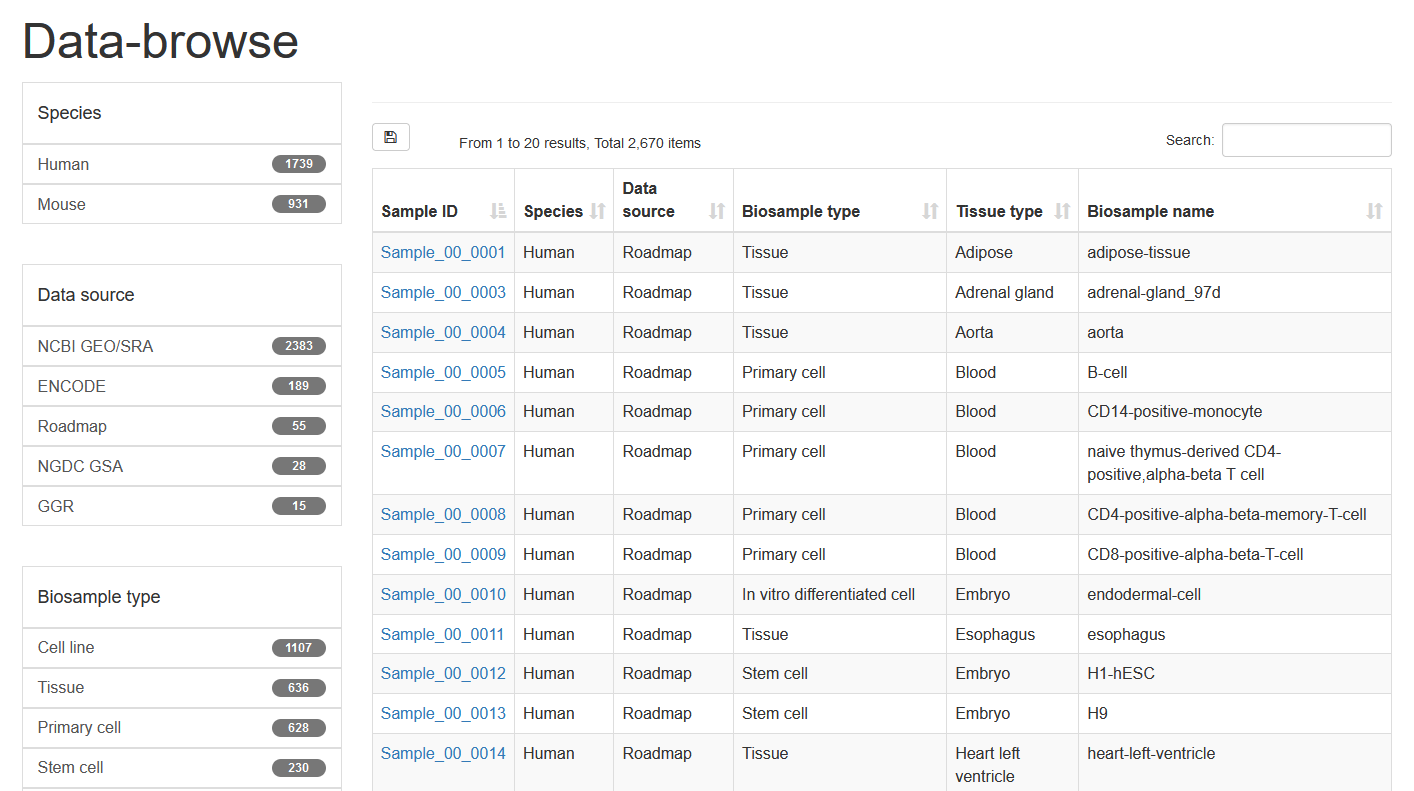
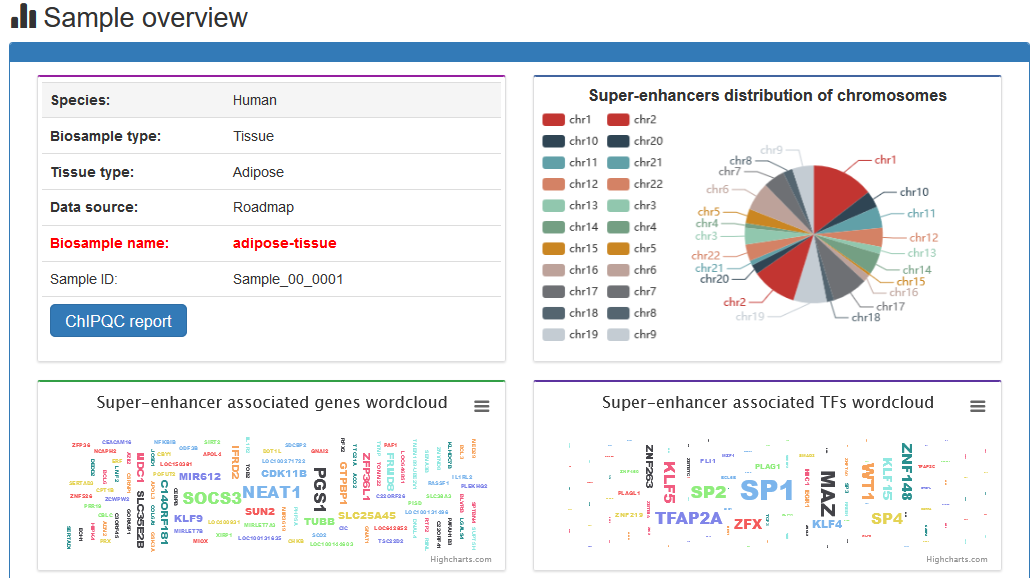
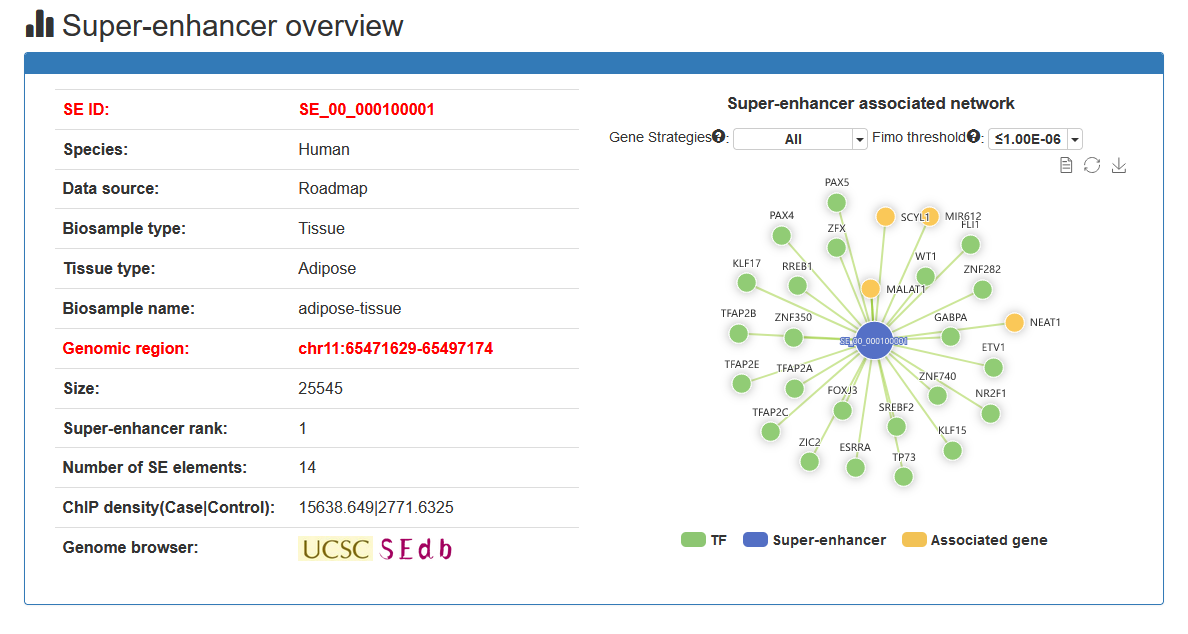
Here, we developed a comprehensive super-enhancer database of human and mouse (SEdb 2.0, http://www.licpathway.net/sedb) that aimed to provide a large number of available resources on human and mouse super-enhancers. The database was annotated with potential functions of super-enhancers in the TF-gene regulation. The current version of SEdb documented a total of 1,717,744 SEs from 2,670 samples including 1,739 human samples and 931 mouse samples. Furthermore, SEdb provides detailed genetic and epigenetic annotation information on super-enhancers. Information includes common SNPs, motif changes, expression quantitative trait locus (eQTL), risk SNPs, transcription factor binding sites (TFBSs), CRISPR/Cas9 target sites, DNase I hypersensitivity sites (DHSs), chromatin accessibility regions, methylation sites, chromatin interactions regions and TADs. For in-depth analyses of super-enhancers, SEdb will help elucidate super-enhancer-related functions and find potential biological effects.
 Increases in data from SEdb 1.0 to SEdb 2.0
Increases in data from SEdb 1.0 to SEdb 2.0
 Increases in data from SEdb 1.0 to SEdb 2.0
Increases in data from SEdb 1.0 to SEdb 2.0 number of samples for each tissue type
number of samples for each tissue type
 number of samples for each tissue type
number of samples for each tissue typeNews and Updates
10/16/2017 Database construction
06/11/2018 The database is online
08/02/2018 Add gene-based and organization-based query capabilities
10/17/2018 SEdb is accepted by Nucleic Acids Research
02/03/2019 SEdb v1.01: Delete a sample (SE_00_002), SEdb now contains 541 samples
03/13/2019 SEdb v1.02: Update the download files to provide super-enhancer associated genes
03/14/2019 SEdb v1.03: Update the page of 'Home' and 'Submit'
05/21/2021 SEdb v1.04: Update the page of 'Data-Browse' and 'Download'
03/18/2022 SEdb v2.00: Add 1198 human sample and 931 mouse sample
06/02/2022 SEdb v2.00: Add Search super-enhancers by TF-based, SE-based TF-Gene analysis and Differential-Overlapping-SE analysis
Sister Projects
ENdb
ENdb: an experimentally supported enhancer database for human and mouse
SEanalysis
SEanalysis: a web tool for super-enhancer associated regulatory analysis
LncSEA
LncSEA: a comprehensive human lncRNA sets resource and enrichment analysis platform
ATACdb
ATACdb: A comprehensive human chromatin accessibility database
KnockTF
KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of transcription
factors
TRCirc
TRCirc: a resource for transcriptional regulation information of circRNAs
TRlnc
TRlnc: a comprehensive database of human transcriptional regulation of lncRNAs
VARAdb
VARAdb: a variation annotation database for human
Contact us
Principal Investigator: Chunquan Li, Ph.D.
The First Affiliated Hospital, Hengyang Medical School, University of South China
Email: lcqbio@163.com
SEdb 1.0 link is http://www.licpathway.net/sedbv1/
Publication
Wang Y, Song C, Zhao J, Zhang Y, Zhao X, Feng C, Zhang G, Zhu J, Wang F, Qian F, Zhou L, Zhang J, Bai X, Ai B, Liu X, Wang Q, Li C. SEdb 2.0: a comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 2022 Nov 1:gkac968. doi: https://doi.org/10.1093/nar/gky1025. PMID: 36318264.