Welcome to KnockTF
a comprehensive human gene expression profile database with knockdown/knockout of transcription factors
What is KnockTF?
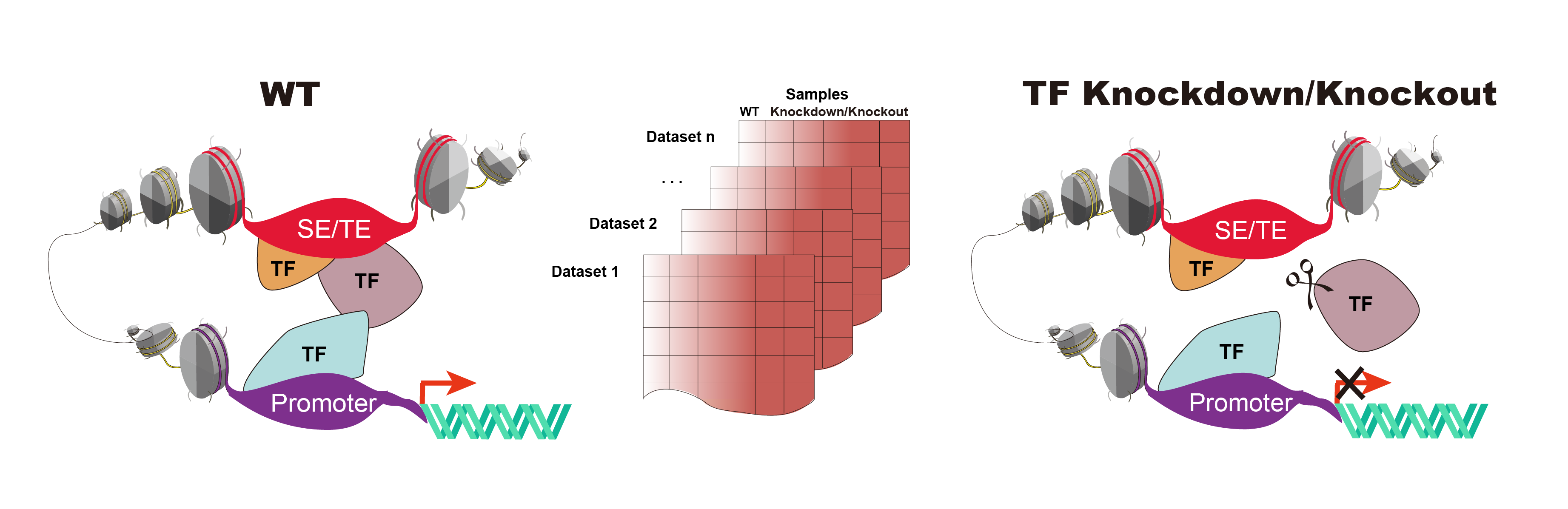
The current version of KnockTF has 570 manually curated RNA-seq and microarray datasets associated with 308 TFs disrupted by different knockdown/knockout techniques and across multiple tissue/cell types. KnockTF not only provides comprehensive gene expression information about target genes of TFs of interest, but also collects upstream pathway information of TFs and various functional annotation and analysis results of downstream target genes, including GSEA, GO enrichment, KEGG pathway enrichment, hierarchical clustering analysis and differentially expressed analysis. KnockTF further provides details about TFs binding to promoters, super-enhancers and typical enhancers of target genes. In addition, a TF-differentially expressed gene network is constructed and used to perform network analyses for gene sets of interest, such as subnetwork location, topological analysis and hypergeometric enrichment. KnockTF will help elucidate TF-related functions and tap potential biological effects.
Contact us
Principal Investigator: Chunquan Li, Ph.D.
School of Medical Informatics,Daqing Campus Harbin Medical University 39 Xinyang Road, Daqing 163319, China
Phone: 86-459-8153035
Fax: 86-459-8153035
Email: lcqbio@163.com
We welcome researchers from all over the world to provide valuable advice for KnockTF, and make KnockTF more and more perfect.
SEdb
SEdb: the comprehensive human Super-Enhancer database
ENdb
ENdb: an experimentally supported enhancer database for human and mouse
SEanalysis
SEanalysis: a web tool for super-enhancer associated regulatory analysis
ATACdb
ATACdb: a comprehensive human chromatin accessibility database
LncSEA
LncSEA: a comprehensive human lncRNA sets resource and enrichment analysis platform.
TRCirc
TRCirc: a resource for transcriptional regulation information of circRNAs
TRlnc
TRlnc: a comprehensive database of human transcriptional regulation of lncRNAs
VARAdb
VARAdb: a variation annotation database for human
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)