Help&FAQs
1. What Information is Available in the KnockTF?
2. What Datasets are Included in the KnockTF?
3. Database Content and Construction
4. How to Use the KnockTF?
4.1 Browse
4.2 Search
4.3 Analysis
4.3.1 Subnetwork Analysis
4.3.2 TF Enrichment
4.4 Download
4.5 Statistics
5. Frequently Asked Questions
6. Development Environment
1. What Information is Available in the KnockTF?
Transcription factors (TFs) and their target genes play significant roles in various human diseases and biological processes. Gene expression profile analysis technique before/after knockdown/knockout has become one of the most important strategies for obtaining target genes of TFs and exploring TF functions. More and more human gene expression profile datasets of TF knockdown/knockout are accumulating rapidly, which promotes the urgent need to comprehensively and effectively collect and process these data.
Here, we develop a comprehensive human gene expression profile database with TF knockdown and knockout. KnockTF (http://www.licpathway.net/KnockTF/index.html) provides a large number of available resources of human gene expression profile datasets associated with TF knockdown/knockout and annotates TFs and their target genes in a tissue/cell type-specific manner.
Here, we develop a comprehensive human gene expression profile database with TF knockdown and knockout. KnockTF (http://www.licpathway.net/KnockTF/index.html) provides a large number of available resources of human gene expression profile datasets associated with TF knockdown/knockout and annotates TFs and their target genes in a tissue/cell type-specific manner.
2. What Datasets are Included in the KnockTF?
The current version of KnockTF has 570 manually curated RNA-seq and microarray datasets associated with 308 TFs disrupted by different knockdown/knockout techniques and across multiple tissue/cell types. KnockTF provides a conveniently, user-friendly interface for querying, browsing, analyzing and downloading detailed information about gene expression profile datasets of TF knockdown/knockout.
For more detailed statistics, please see the "Statistics" page.
For more detailed statistics, please see the "Statistics" page.
3. Database Content and Construction
KnockTF not only provides comprehensive gene expression information about target genes before/after knockdown/knockout of TFs of interest, but also collects upstream pathway information of TFs and various functional annotation and analysis results of downstream target genes, including Gene Set Enrichment Analysis, Gene ontology enrichment, KEGG pathway enrichment, hierarchical clustering analysis and differentially expressed analysis. Furthermore, KnockTF provides detailed information about TFs binding to promoters, super-enhancers and typical enhancers of target genes. In addition, KnockTF constructs a TF-differentially expressed gene network and performs network analyses for gene sets of interest, such as subnetwork location, topological analysis and hypergeometric enrichment.
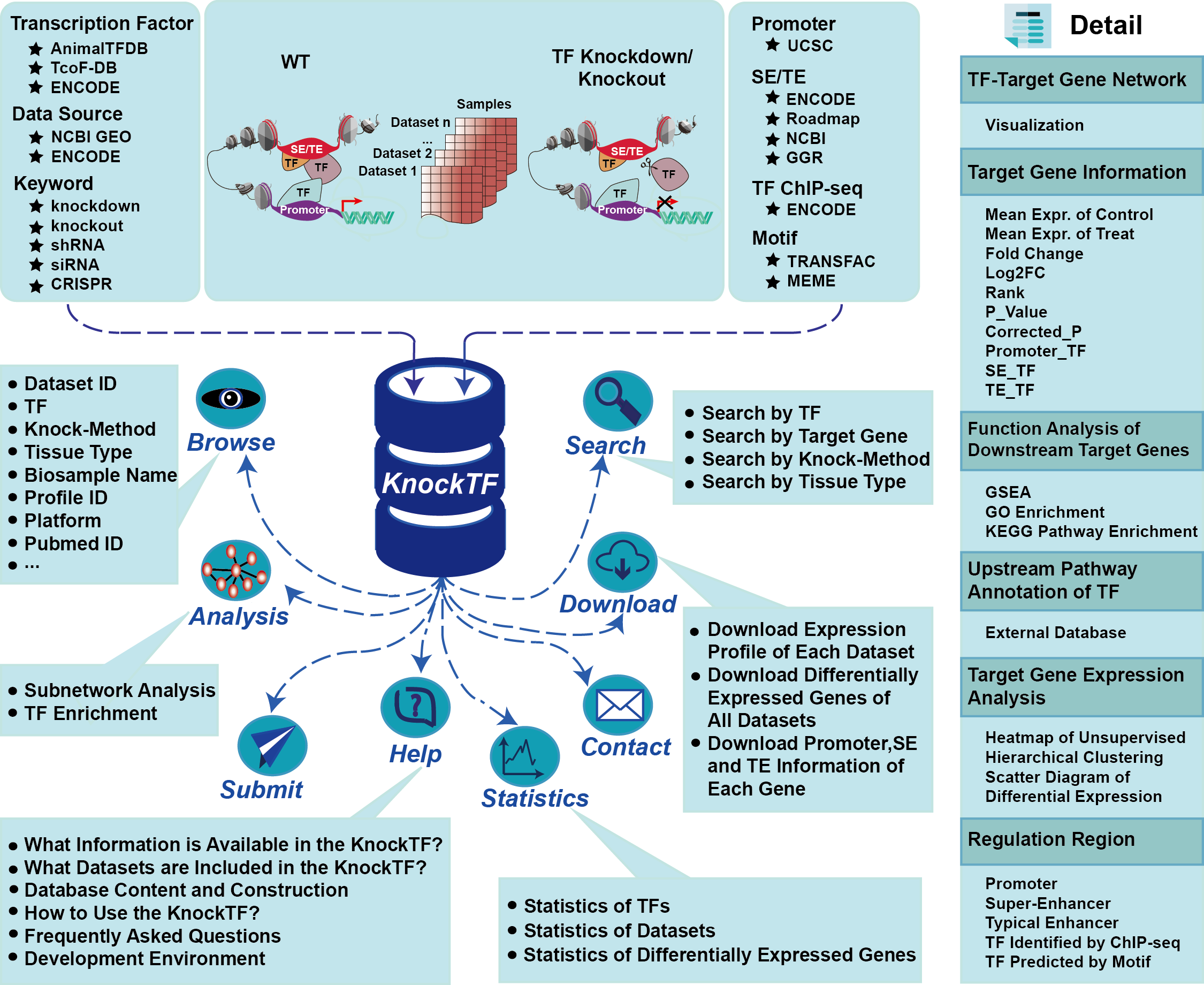

4.How to Use the KnockTF?
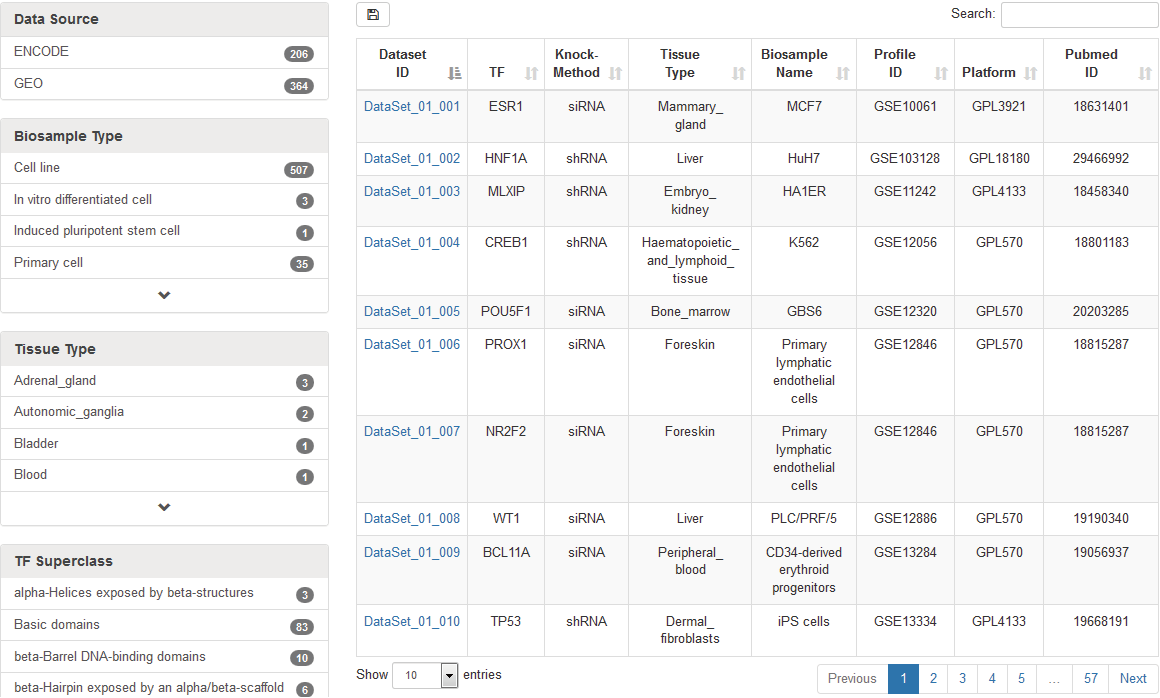
The “Browse” page is organized as an interactive table for quickly searching for TF knockdown/knockout datasets and customizing filters using “Data Source”, “Biosample Type”, “Tissue Type”, “TF Superclass” and “TF”. Users can click the “Show entries” in a dropdown menu to change the number of records displayed per page. To view details of a given TF knockdown/knockout dataset, users click on “Dataset ID”.

KnockTF supports four different searching modes: “Search by TF”, “Search by Target Gene”, “Search by Knock-Method” and “Search by Tissue Type”. Brief search results are presented as a table in the result page.
As an example, users can click Search→Search by TF→CREB1, the database will automatically display TF Class and TF Superclass: Basic leucine zipper factors (bZIP) and Basic domains. Search results are shown on the next page, including Dataset ID, TF, Knock-Method, Biosample Type, Tissue Type, Biosample Name, Profile ID, Platform, The Number of Control, The Number of Treat and Pubmed ID. Users can click the “Dataset ID” to view details about each TF knockdown/knockout dataset.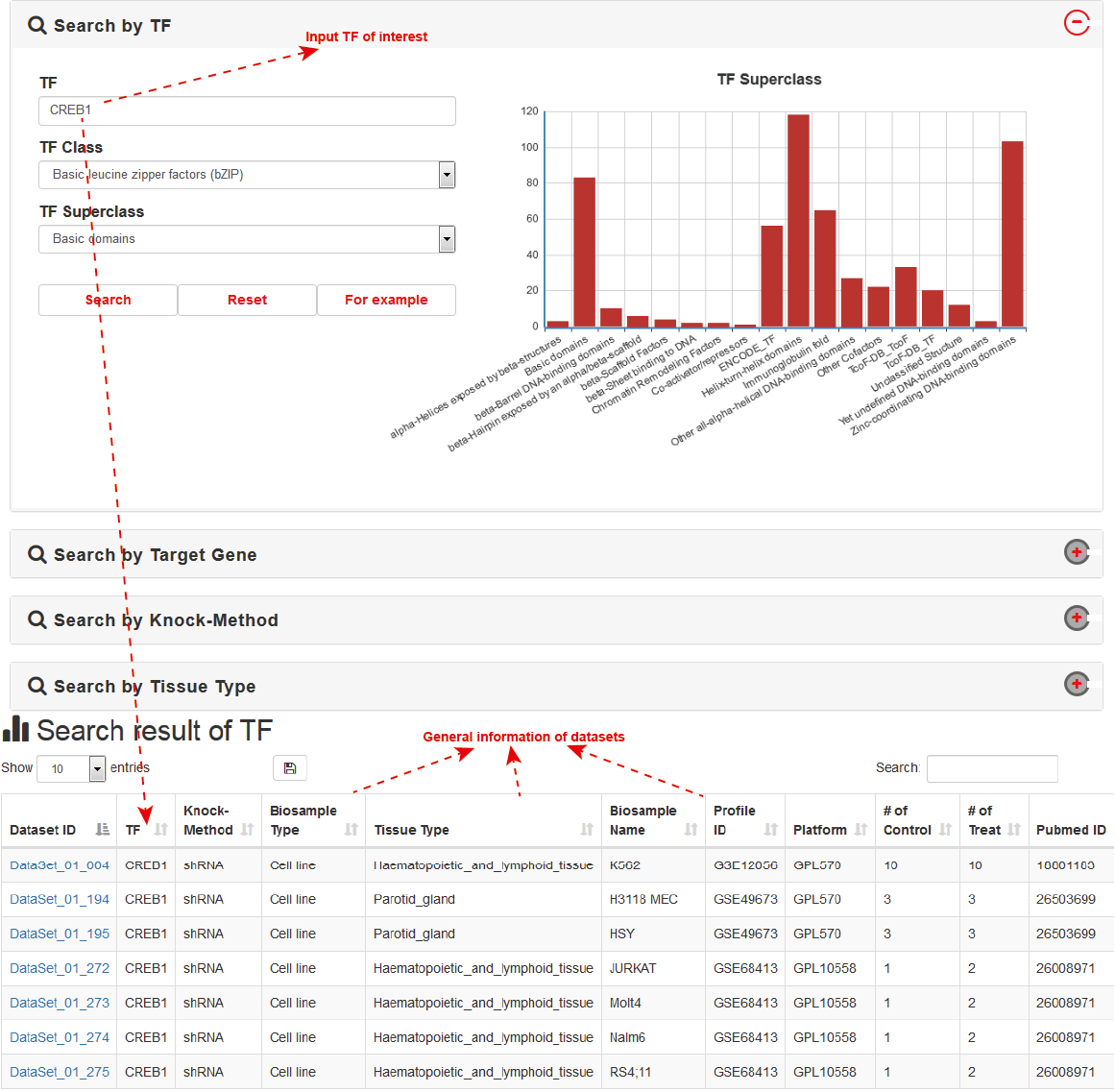
As an example, users can click Search→Search by TF→CREB1, the database will automatically display TF Class and TF Superclass: Basic leucine zipper factors (bZIP) and Basic domains. Search results are shown on the next page, including Dataset ID, TF, Knock-Method, Biosample Type, Tissue Type, Biosample Name, Profile ID, Platform, The Number of Control, The Number of Treat and Pubmed ID. Users can click the “Dataset ID” to view details about each TF knockdown/knockout dataset.

The “Search result of Dataset” page presents TF overview, TF-target gene network, target gene information, upstream pathway annotation of TF and various functional annotation and analysis results of downstream target genes, including Gene Set Enrichment Analysis (GSEA), Gene ontology (GO) enrichment, KEGG pathway enrichment, hierarchical clustering analysis and differentially expressed analysis.


Users can further click a target gene name, eg. “AIM1”, to view detailed descriptions of a gene in new page, including gene overview, differentially expressed (FC≥3/2 & FC≤2/3) target gene-TF pairs, annotation of TF binding regions of target gene of interest and gene expression atlas from different data sources.
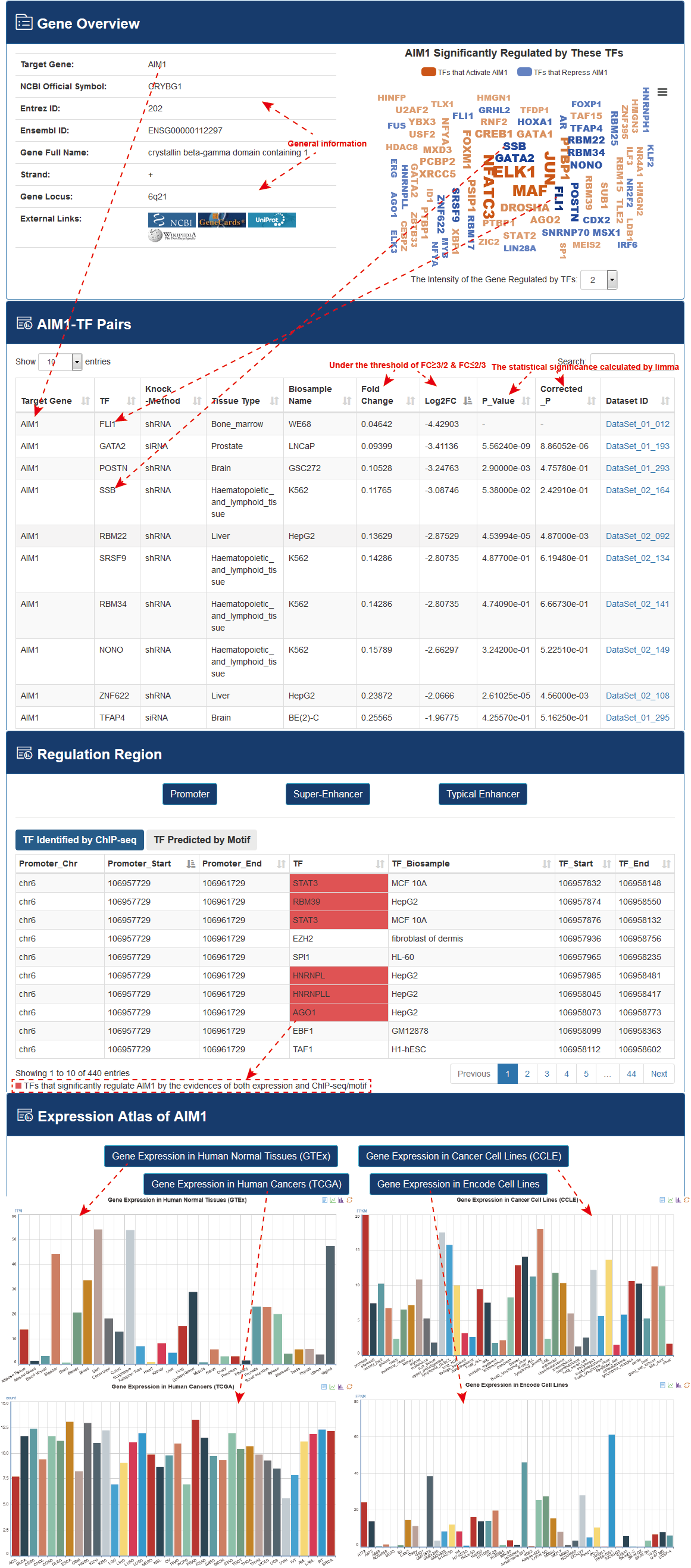

KnockTF constructed a TF-differentially expressed gene (DEG) network, as follows. First, for each TF knockdown/knockout dataset, we extracted DEGs under the threshold of FC≥3/2 & FC≤2/3 and formed TF-DEG pairs that were ranked based on significant levels of DEGs. Second, we combined all TF-DEG pairs for the 570 TF knockdown/knockout datasets. If a TF-DEG pair appeared multiple times in different TF knockdown/knockout datasets, we removed duplications and retained its minimum rank. Then, we reordered all nonredundant TF-DEG pairs and constructed a TF-DEG network with TFs and their DEGs as nodes and TF-DEG pairs as edges. The rank of TF-DEG pairs represented the importance of the regulatory intensity of TFs on target genes. TF-target relationships supported by the ChIP-seq data were also marked and recorded for TF-DEG pairs. Topological features such as degree, betweenness and closeness of all nodes in the TF-DEG network were computed.
4.3.1 Subnetwork Analysis
Users can submit a gene list to locate a transcriptional regulatory subnetwork. The subnetwork consists of submitted genes and their one-step neighbors within TF-DEG network. TF-target gene relationships supported by the ChIP-seq data have bold edges in the subnetwork. Users can choose subnetwork size displays by filtering the number of the most important TF-DEG pairs. KnockTF also provides topological features of subnetwork genes including degree, betweenness and closeness.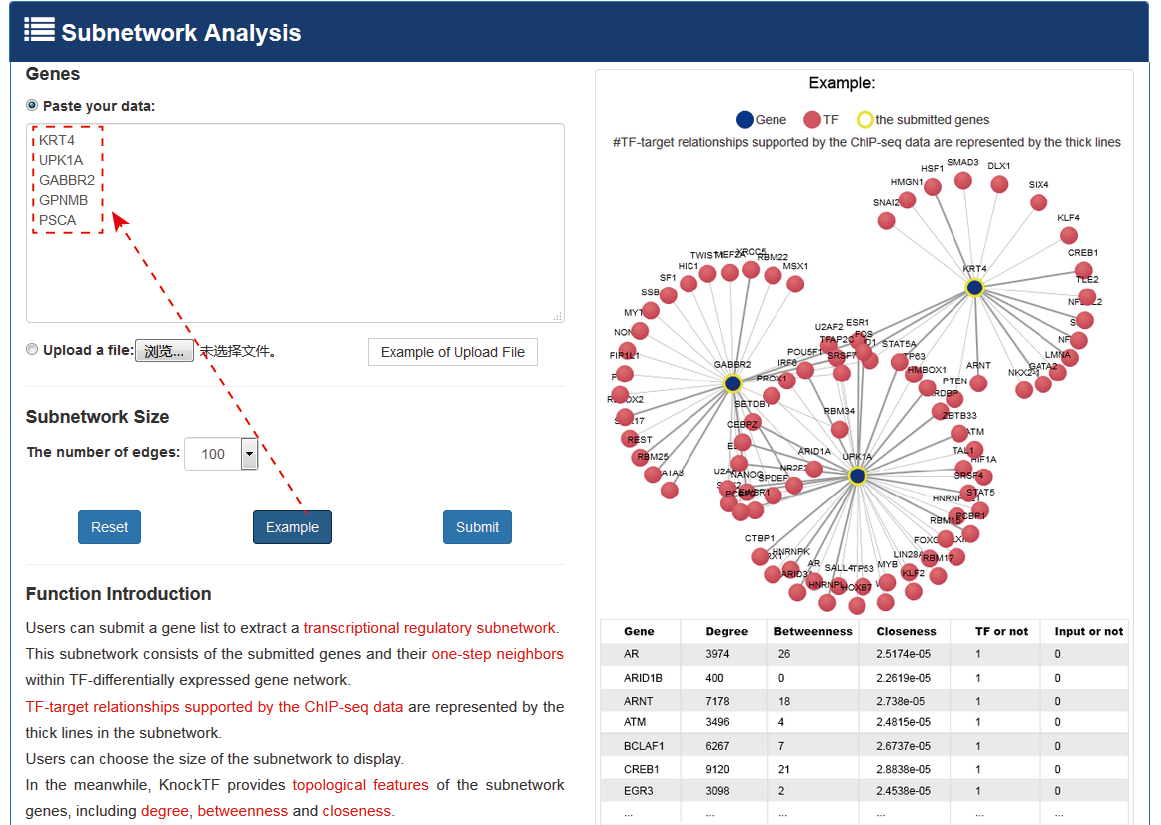
4.3.2 TF Enrichment
Users can submit a gene list and set (FDR-adjusted) P-value for TF enrichment. KnockTF maps submitted genes to the TF-DEG network and performs hypergeometric test between submitted genes and all DEGs regulated by each TF. The TFs under the threshold of (FDR-adjusted) P-value user sets are considered the most important TFs that significantly regulate the submitted genes.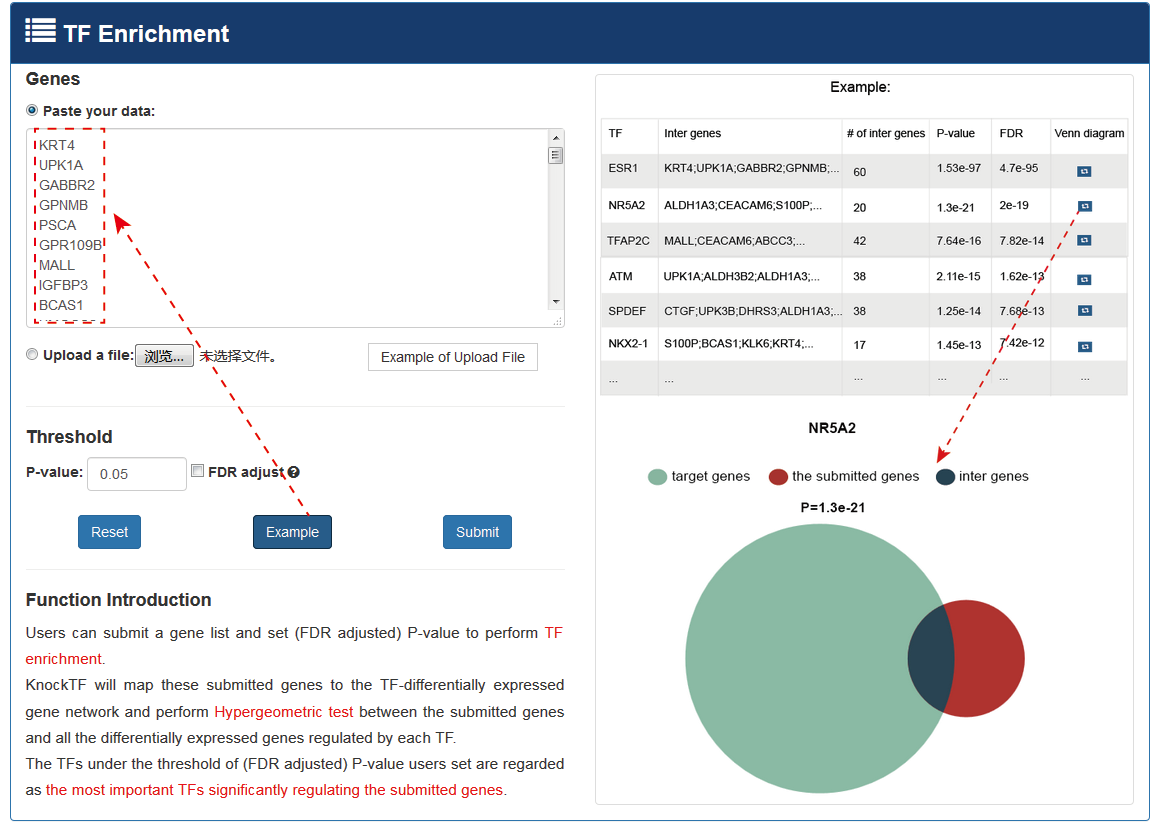
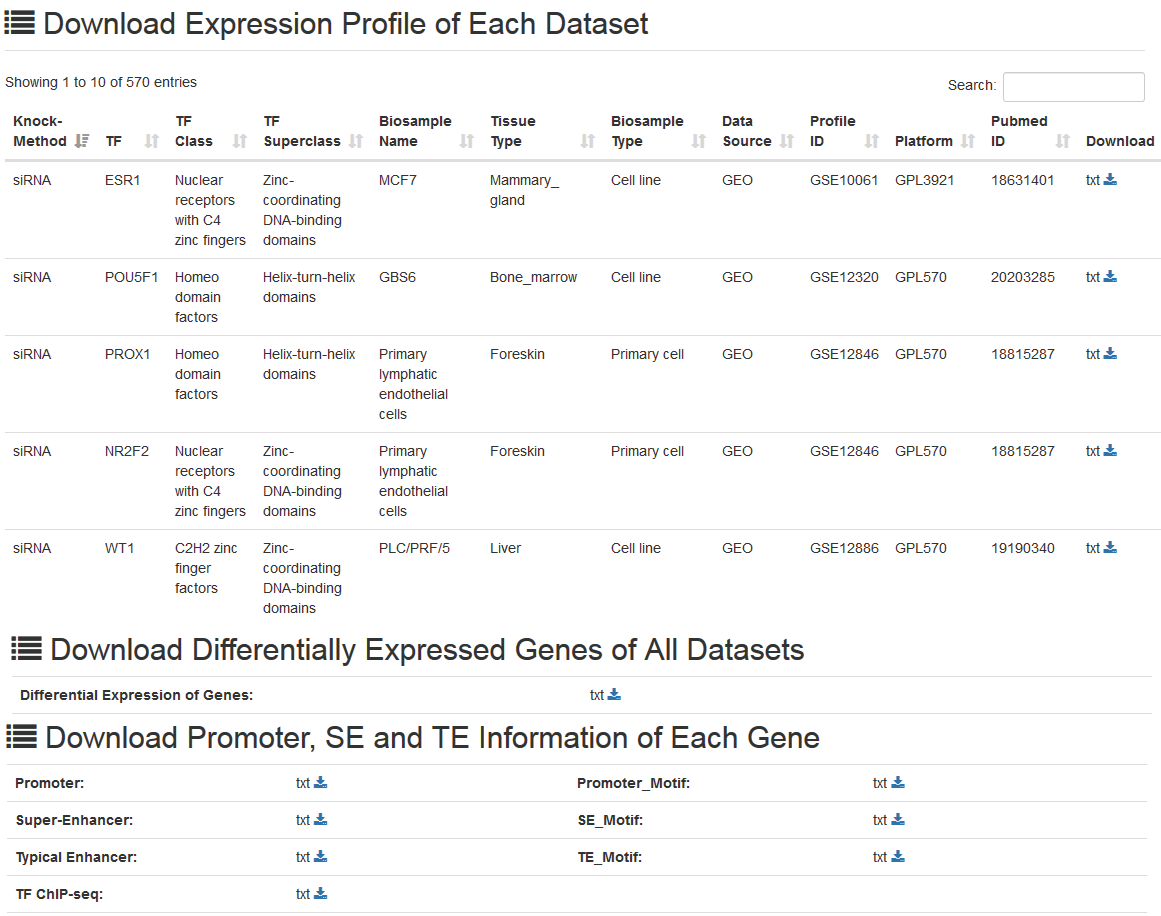
Gene expression profile of each dataset, differential expression information of genes, the promoter/super-enhancer/typical enhancer regions and corresponding TF binding information of target genes are provided for downloading in the “Download” page. In addition, KnockTF supports export of query results for each search result page.

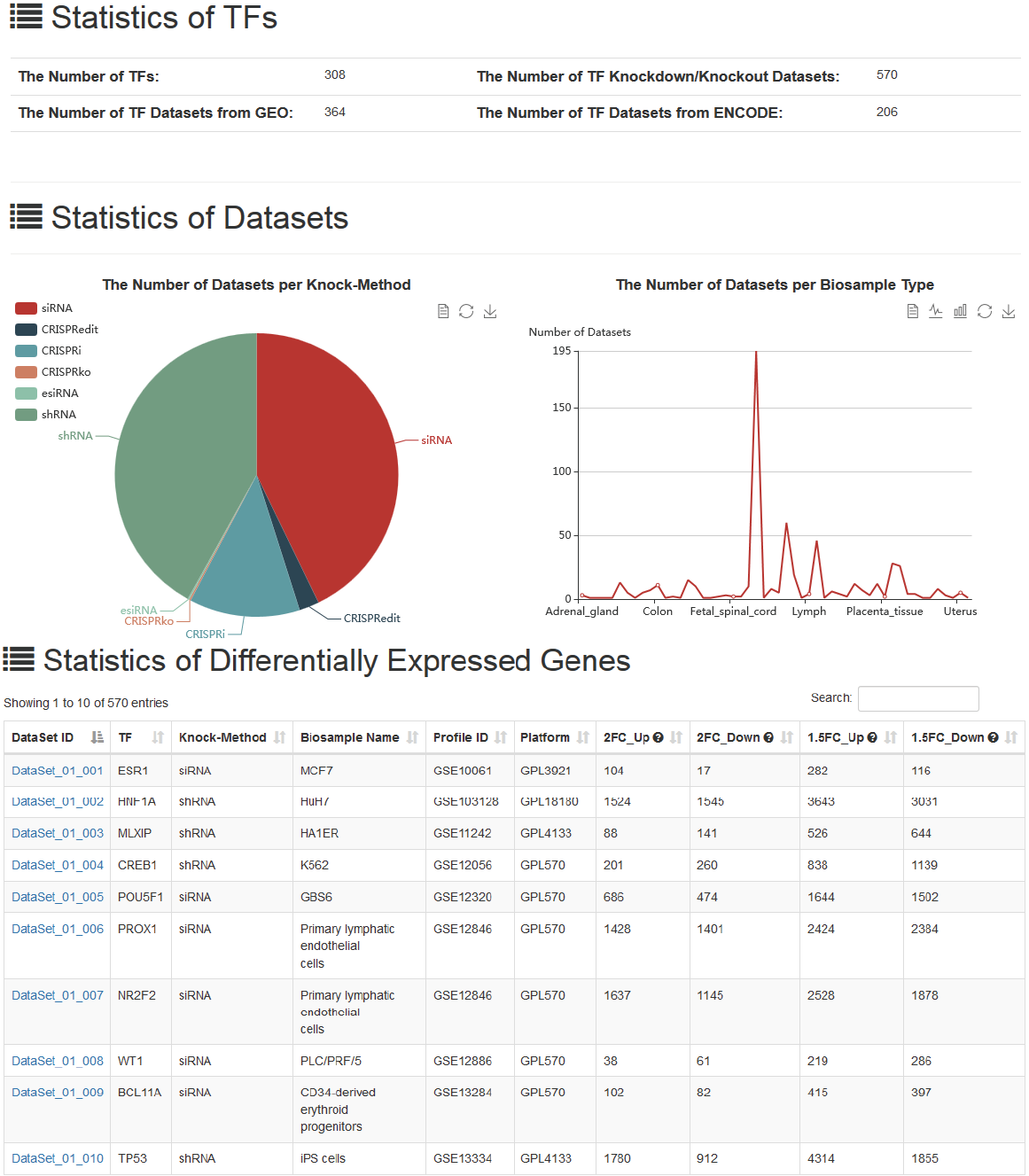
The datasets are respectively classified by different knock-methods and biosample types in the “Statistics” page. The number of DEGs in each TF knockdown/knockout dataset is also provided.

5. Frequently Asked Questions
5.1 Why are showing no results available somewhere?
Reply: Because there is no corresponding function annotation or no significant annotation results for this TF/gene.5.2 Why the same TF disrupted by the same knockdown/knockout technique appears in different datasets?
Reply: Because these datasets may be from different profiles, different platforms or different biosamples. In KnockTF, there are two data sources. If data source is GEO, five conditions are used to uniquely determine a dataset, including TF, knockdown/knockout technique, biosample name, series (GSE) and platform (GPL); if data source is ENCODE, four conditions are used to uniquely determine a dataset, including TF, knockdown/knockout technique, biosample name and experiment accession.5.3 Which kinds of species are stored in the database?
Reply: The current version of KnockTF stores the most abundant human gene expression profile datasets of TF knockdown/knockout. There may be a lot of TF knockdown/knockout data about other species in other data sources. In the next version of KnockTF, we will manually curate more TF knockdown/knockout data with more species and enrich the kinds of species information.5.4 Why might web pages load slowly?
Reply: KnockTF has advanced storage technology and sufficient bandwidth to meet the needs of most users for the speed of web page loading. However, it is not excluded that few users have poor user experience due to network reasons.
6.Development Environment
The current version of KnockTF was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). PHP 7.0 (http://www.php.net) was used for server-side scripting. The interactive interface was designed and built using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). ECharts (https://www.echartsjs.com/) and Highcharts (https://www.highcharts.com.cn/) were used as a graphical visualization framework. We recommend to use a modern web browser that supports the HTML5 standard, such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The knockTF database is freely available to the research community using the web link (http://www.licpathway.net/KnockTF/index.html). Users are not required to register or login to access features in the database.
The knockTF database is freely available to the research community using the web link (http://www.licpathway.net/KnockTF/index.html). Users are not required to register or login to access features in the database.