VARAdb!v1.0
A variation annotation database for human
What is VARAdb?
Here, we developed a comprehensive human variation annotation database (VARAdb,
http://www.licpathway.net/VARAdb), which aims to provide a large number of variations and annotate their
potential roles with a large amount of regulatory information.
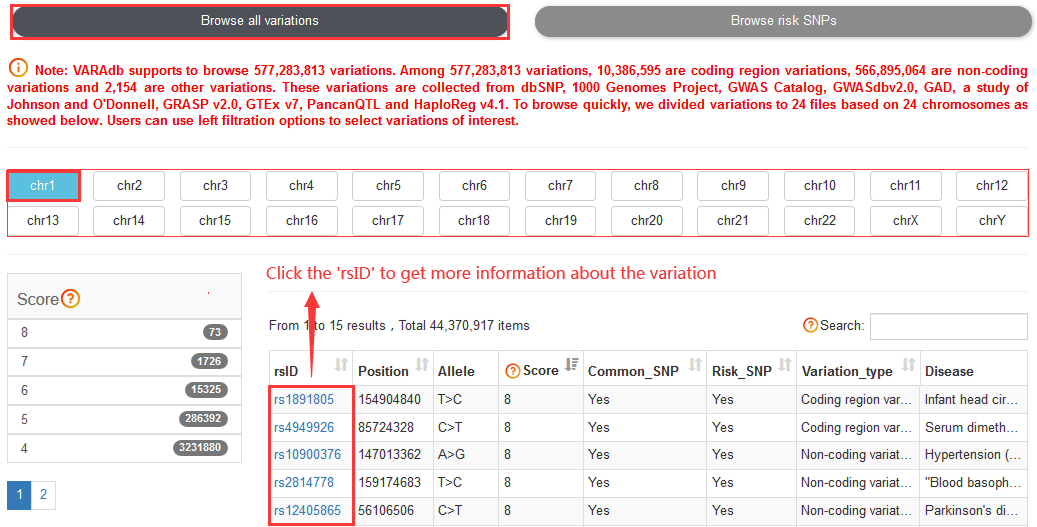
The current version of VARAdb cataloged a total of 577,283,813 variations and
provided five annotation sections including ‘Variation
information’, ‘Regulatory
information’, ‘Related genes’, ‘Chromatin
accessibility’ and ‘Chromatin interaction’, with significantly more information than similar databases. The information includes motif changes, risk SNPs, LD SNPs,
eQTLs, clinical variant-drug-gene pairs, sequence conservation, somatic mutations, enhancers, super
enhancers, promoters, TFs, ChromHMM states, histone modifications, ATAC accessible regions and chromatin
interactions from Hi-C and ChIA-PET.
Importantly, we considered two types of
variation related genes: 1)
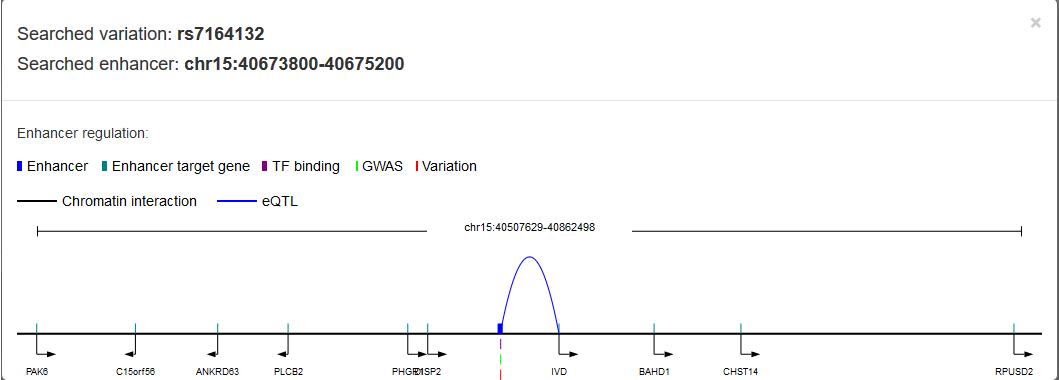
variation that sets in enhancer may associate with enhancer target genes predicted by Lasso method; 2)
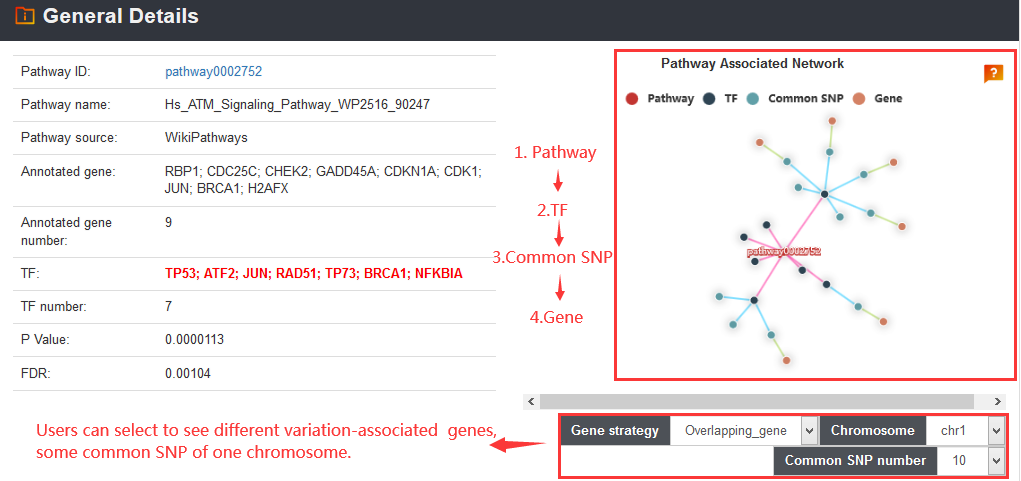
variation related genes based on distance. In addition, VARAdb can
prioritize variations based on score, annotate novel variants and perform pathway downstream analysis
conveniently. Together, VARAdb is a user-friendly database to query, browse and visualize variations of
interest. We believe VARAdb will help obtain perspectives on the regulation of variations in complex
diseases.
Collection of variations
We have not only collected the variation from dbSNP but also multiple other resources. Notably, 577,098,938 SNVs were collected from dbSNP v151 and 79,482,384 common SNPs were collected from the 1000 Genomes Project. Each common SNP from the 1000 Genomes Project has at least one 1000 Genomes population with a minor allele of frequency ≥ 1%. Millions of LD SNPs of five super-populations (4,477,132 from African; 4,548,152 from Ad Mixed American; 3,693,208 from East Asian; 4,011,947 from European; and 3,838,175 from South Asian) were calculated using phased genotype information accompanying the 1000 Genomes Project phase 3. In addition, we integrated 1,515,001 risk SNPs from the GWAS Catalog, GWASdbv2.0, GAD, Johnson and O'Donnell, and GRASP v2.0. We also obtained 3,998,301 eQTLs from GTEx v7, PancanQTL, and HaploReg v4.1.
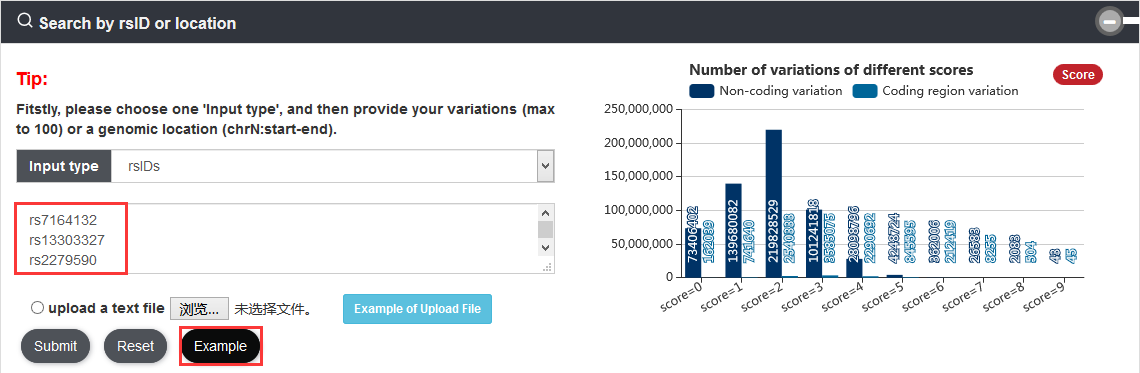
Note: We calculated score of the variation that means how many categories the variation associated with. Each variation is scored based on its annotated records on nine annotation categories: risk SNP, eQTL, motif change, conservation, enhancer and super enhancer, promoter, TF binding, ATAC accessible region and Hi-C.

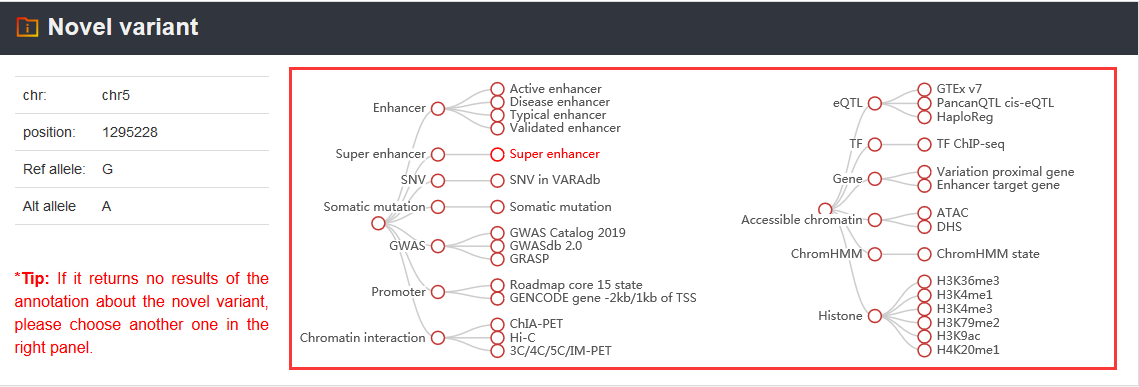
Note: VARAdb provides 5 annotation sections for the cataloged and novel variations. These sections are shown as below.
Regulatory infomation






Related genes

Chromatin accessibility

Chromatin interaction

Variation information




Statistics Table
| Variations | 577,283,813 |
| Enhancer sources | 8 |
| Enhancer number | 7,841,333 |
| Enhancer-Gene pairs | 5,880,825 |
| Enhancer-Gene pair sources | 3 |
| TF ChIP-seq sources | 5 |
| TF ChIP-seq samples | 7734 |
| TF number | 1261 |
| Promoter sources | 2 |
| Promoter number | 6203292 |
| Pathway sources | 10 |
| Pathway number | 2880 |
Details please see Statistics
Note:Coding region variations account for ~1.79% of 577,283,813 variations. Among all variations, 10,386,595 are coding region variations, 566,895,064 are non-coding variations and 2,154 are other variations.
Sister Projects
SEdb
SEdb: The comprehensive human Super-Enhancer databasebr
SEanalysis
SEanalysis: a web tool for super-enhancer associated regulatory analysis
KnockTF
KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of
transcription
factors
ENdb
ENdb: An experimentally supported enhancer database for human and mouse
News and Updates
2020.10 VARAdb is accepted by Nucleic Acids Research
2020.05 Browse page is designed
2019.12 The database is online
2019.09 Database construction
For publication of results please cite the following article
Pan Q, Liu YJ, Bai XF, Han XL, Jiang Y, Ai B, Shi SS, Wang F, Xu MC, Wang YZ, Zhao J, Chen JX, Zhang J, Li XC, Zhu J, Zhang GR, Wang QY, Li CQ. VARAdb: a comprehensive variation annotation database for human. Nucleic Acids Res. 2020 Oct 23:gkaa922. doi: 10.1093/nar/gkaa922. Epub ahead of print. PMID: 33095866.