Help
1.Database content and construction
2.Overall workflow and a self-determined method
3.How to use the ENdb?
4.Explanation of the definitions used by the website
5.Development environment
Database content and construction.The user-friendly interface was provided to search,browse,download and visualize the detailed information.
i. Get the accurate location information of the publications (e.g. the figures, text description of location information or SNP loci).
ii. Follow up the cited experimental procedure publications.
iii. The reference genome version was obtained directly according to the data provided in the publications.
i. The screen resolution is unified and resized the images in the publication as 300%.
ii. The screen pixel values obtained by manual measurement are A, B from the transcription start site respectively. A free software for screen pixel was used to measure the pixel values of A, B and P (http://www.ucbug.com/soft/113483.html).
iii. We obtained the start site of target genes from NCBI, and converted the corresponding version of location information by LiftOver in the UCSC browser, then, the formulas (1) and (2) were used to calculate the enhancer region.
The start position of enhancer = the start site of target gene±(A+W/2)/P*xxxb (1)
The end position of enhancer = the start site of target gene±(B-W/2)/P*xxxb (2)
The '±' represents downstream or upstream in the start site of target gene.
iv. The region of enhancer is converted into hg19 or mm10 version by LiftOver.
The 'Data-Browse' page is organized as an interactive and alphanumerically sortable table that allows users to quickly search for samples and customize filters through 'Species', 'Biosample name', 'Enhancer experiment', and 'Disease'. The 'Show entries' drop-down menu are provided for changing the number of records per page conveniently. Viewing the detailed enhancer information, users can simply search the 'Enhancer ID'.
Users can search enhancers through six paths, including 'Searching by enhancer', 'Searching by target gene', 'Searching by TF', 'Searching by SNP', 'Searching by Tissue', 'Searching by Cell'. What's more, all the information of enhancers can be got through inputting the start position and end position. Users can also submit their own filters. And the format needs keep accordance with our examples.
Users can click 'Details', and then the detailed information of enhancer, including the interaction of enhancer associated netowrk, expression of target genes and SNPs in the enhancer region will be displayed on the next page.
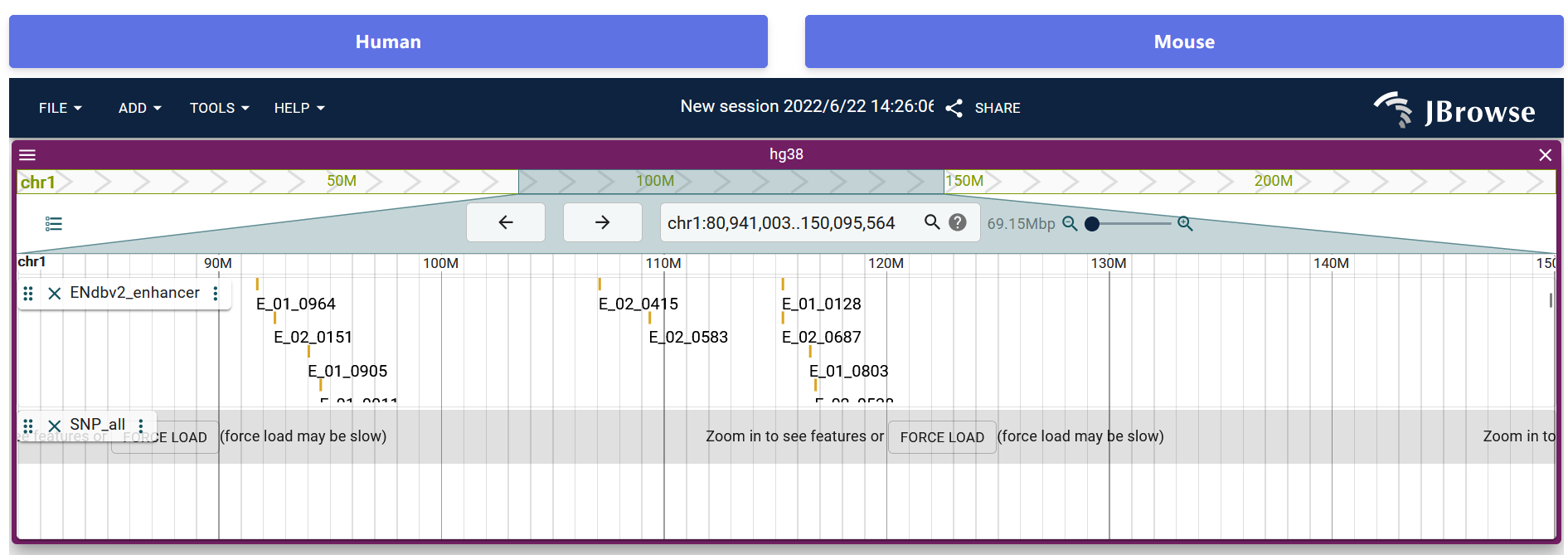
In order to help users to view the proximity information of enhancers in the genome intuitively, we developed a personalized genome browser using JBrowse2 and added a lot of useful tracks. Users see the proximity of enhancers to nearby genes, genome segments, SNPs and TFs.
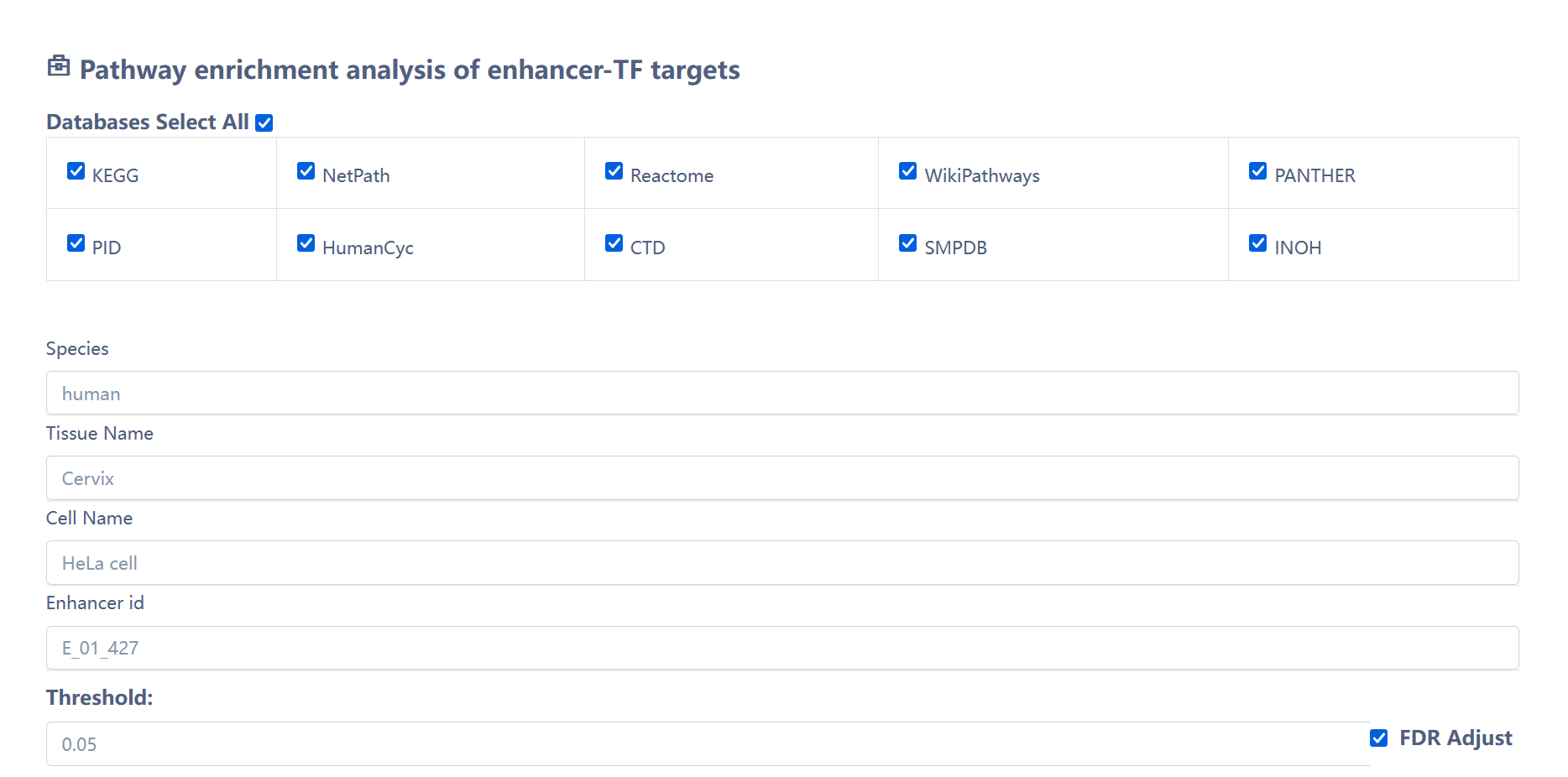
We provide the pathway enrichment analysis function of enhancer related transcription factor target genes. Users can input endb enhancer ID, and the website can automatically identify the related transcription factor target genes for analysis.
If you want to share your data, please fill out the necessary information in the form below and we will add it to ENdb soon. Thank you.
The data of Enhancers and their elements of all samples are provided for download in the 'Download' page. Users can download their files of interest.
| ENdb database contains 24 columns separated by tab: | Description |
|---|---|
| Pubmed ID | PMID of the article. |
| Symbol | The named enhancer in the article. |
| Reference gene | Reference genome version number (hg19,mm10). |
| Chromsome | The chromsome number. |
| Start position | The start position of the enhancer. |
| End position | The end position of the enhancer. |
| Species | Species,including human and mouse. |
| Sample | The specific tissues or cells used in the experiment, other species cannot be explicitly described or described |
| Experiment class( High throughput/Low throughput) | Experimental categories( High throughput/Low throughput). |
| Enhancer experiment | Experiments used to prove the activity of enhancer. |
| Enhancer type | Enhancer type(enhancer/super-enhancer). |
| Experiment description of enhancer target gene | Experimental description of enhancer result. |
| Target gene | The target genes of the enhancer was confirmed by experiments. |
| Disease | Disease name. |
| Enhancer function | The function of enhancers is verified experimentally. |
| Enhancer of the enhancers function | Experiments on the function of enhancers. |
| Enhancer description of enhancers function | Experimental description of the function of enhancers. |
| TF name | The name of the transcription factor. |
| TF experiment | Experiments on the TF of enhancers. |
| SNP symbol | The Literature-supported SNP. |
The current version of ENdb is developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). The server-side scripting is PHP 7.0 (http://www.php.net) . We designed and built the interactive interface using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). We used ECharts (http://echarts.baidu.com) and D3 (https://d3js.org) as a graphical visualization framework, and JBrowse (http://jbrowse.org) is the genome browser framework. We recommend using a modern web browser that supports the HTML5 standard such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The ENdb database is freely available to the research community using the web link (http://www.licpathway.net/ENdb). Users are not required to register or login to access features in the database.
Material disclaimerThe materials and frameworks used by ENdb are shared by the network and do not contain intellectual property infringement. If there is any infringement, please write to us and we will change it in time.