| Sample ID: | Sample_0013 |
| Biosample name: | hPGCLCs induced from primed UCLA2 hESCs |
| Biosample type: | Stem Cell |
| Tissue type: | Embryo |
| Cancer type: | Normal |
| Region number: | 65085 |
| Length: | 41953.36 kb |
| GEO/SRA ID: | SRR7942734 |
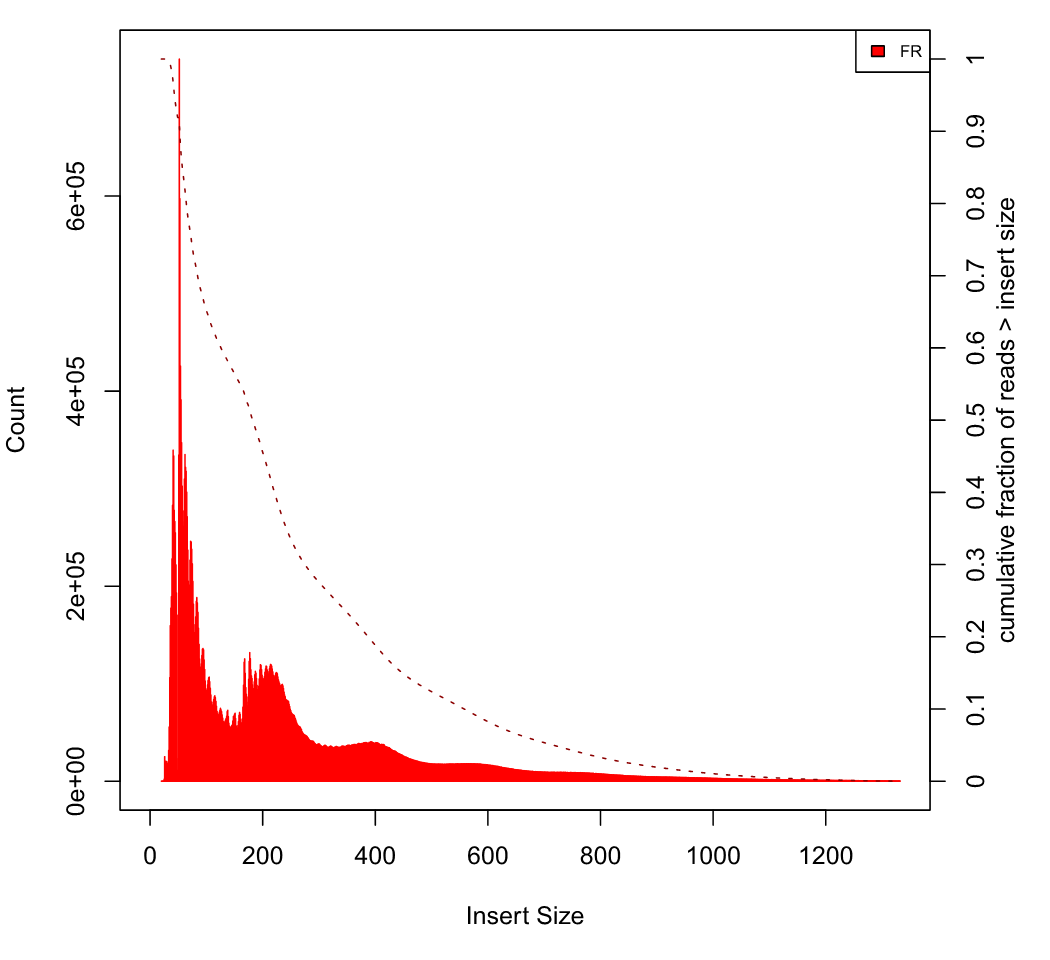
| Mean insert size: | 242.094 |
| Standard deviation: | 219.885 |
| TSS enrichment score: | 22.49390 |
| FRiP: | 0.14054 |
| Diasgnostic plot | |
 |
|
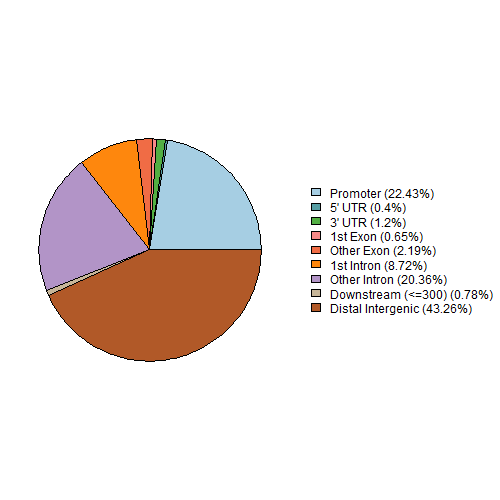
| Plot A: a pie chart of annotated genomic features. |
| Note A: the proportion of annotating genomic region (Promoter, 5'UTR, 3'UTR, Exon, Intron, Downstream and Intergenic). |
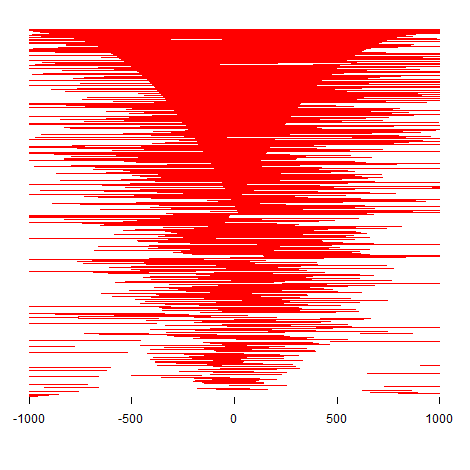
| Plot B: a heatmap of peaks binding to TSS region. |
| Note B: the profile of ATAC peaks binding to TSS region (+/- 1kb). |
| Read mapping: | bowtie2(v2.2.5) -p 10 -X 2000 -x -S |
| Duplicates Removing: | picard MarkDuplicates REMOVE_DUPLICATES=true ASSUME_SORTED=true |
| Peak calling: | macs2(2.1.2) --broad--SPMR --nomodel --extsize 200 -q 0.01 |
| Trimming the adapter: | trim_galore --phred33 --stringency 3 --length 20 |