Accessible chromatin is a highly informative structural feature for identifying regulatory elements, which provides a large amount of information about transcriptional activity and gene regulatory mechanisms, and is closely associated with various biological processes and human diseases. Human ATAC-seq datasets are accumulating rapidly, it is necessary to collect and process these datasets. More importantly, a large number of researches showed that accessible chromatin regions, as well as the related SNPs and TFs, have a strong influence on human diseases and biological processes.
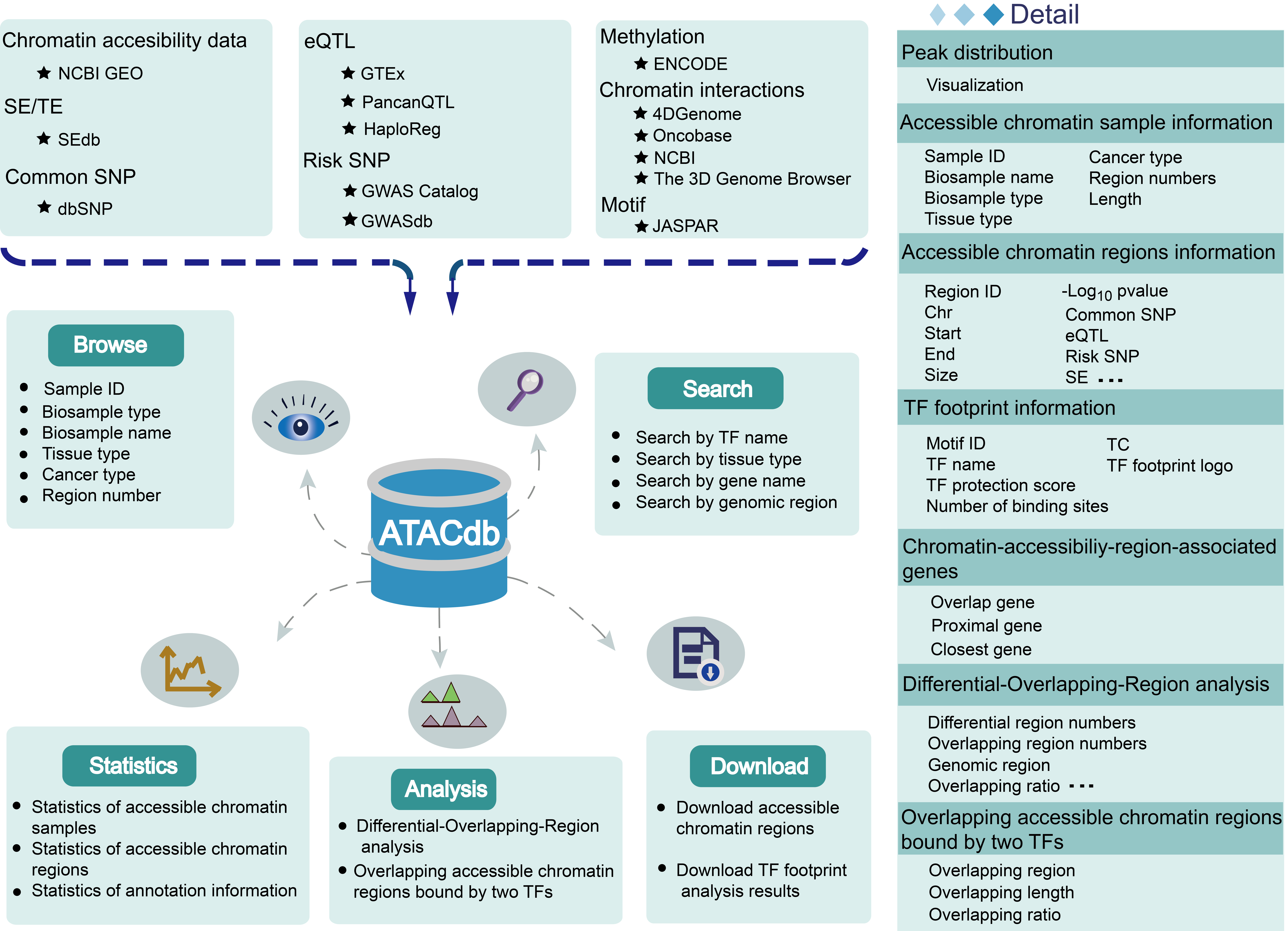
Here, we developed a comprehensive human chromatin accessibility database (ATACdb, http://www.licpathway.net/ATACdb), which aims to provide a large number of available resources on human chromatin accessibility data. ATACdb provides a quality assurance process including four quality control (QC) metrics. ATACdb provides other detailed (epi) genetic annotation information in chromatin accessibility regions, including super-enhancers, typical enhancers, TFs, common SNPs, risk SNPs, eQTLs, LD SNPs, DNA methylation sites, 3D chromatin interactions and TADs. ATACdb provides accurate inference of TF footprint within chromatin accessibility regions. ATACdb is a powerful platform that provides the most comprehensive accessible chromatin data, quality control report, TF footprint and various other annotation information for users.
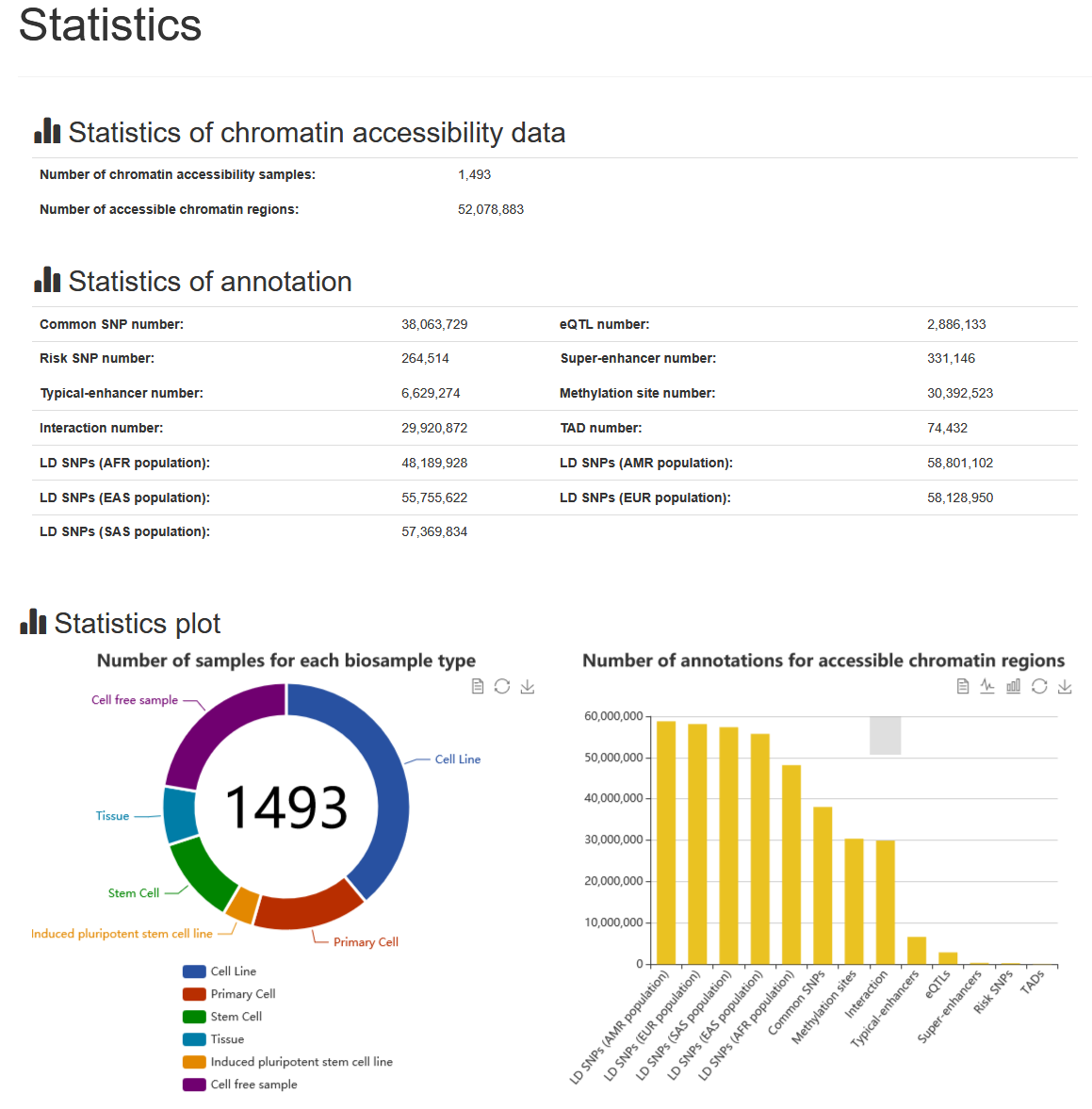
The current version of ATACdb documented a total of 52,078,883 regions from over 1400 chromatin accessibility ATAC-seq samples. These samples have been manually curated and classified from NCBI GEO/SRA. Accessibile chromatin regions were identified by using a unified system environment and software parameters. ATACdb provides a conveniently user-friendly interface to query, browse, analyze and download accessible chromatin regions and their related annotation information.
For more detailed statistics, please see the "Statistics" page.
ATACdb provides the comprehensive chromatin accessibility regions about human ATAC-seq data. ATACdb also provides quality control report and other detailed (epi) genetic information, including super-enhancers, typical enhancers, common SNPs, risk SNPs, eQTLs, LD SNPs, methylation sites, 3D chromatin interactions and TADs. Furthermore, ATACdb includes analytical tools and personalized genome browser to discover potential biological effects of accessible chromatin regions.
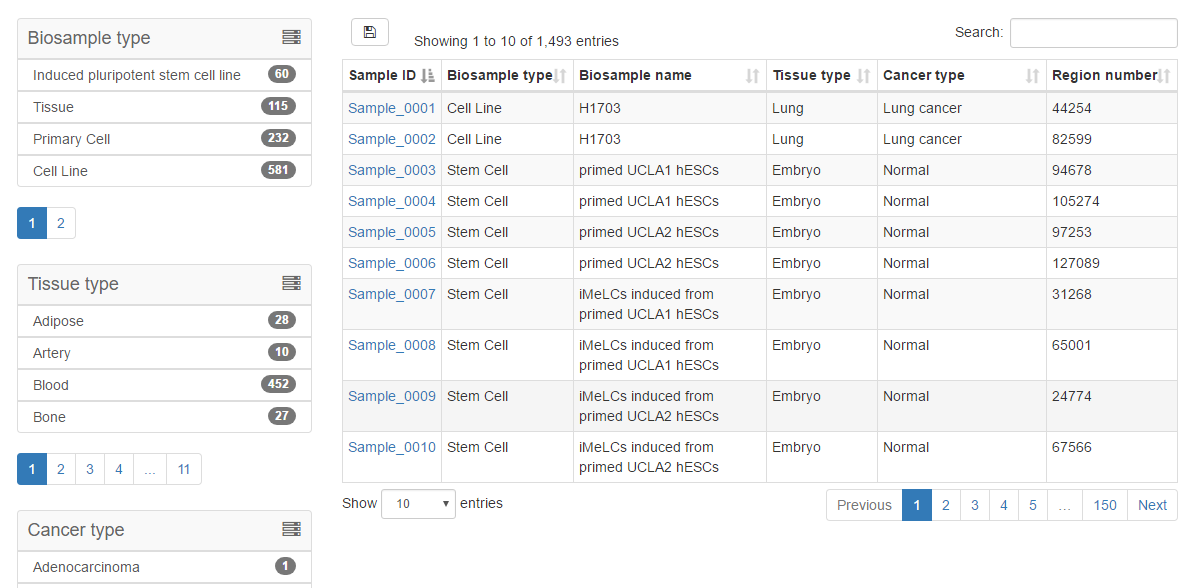
The “Browse” page is organized as an interactive table for quickly searching for samples and customizing filters using “Biosample Type”, “Biosample Name”, “Tissue Type” and “Cancer Type”. Users can click the “Show entries” in a dropdown menu to change the number of records displayed per page. Users can click on “Sample ID” to view details of chromatin accessibility data for a given sample.
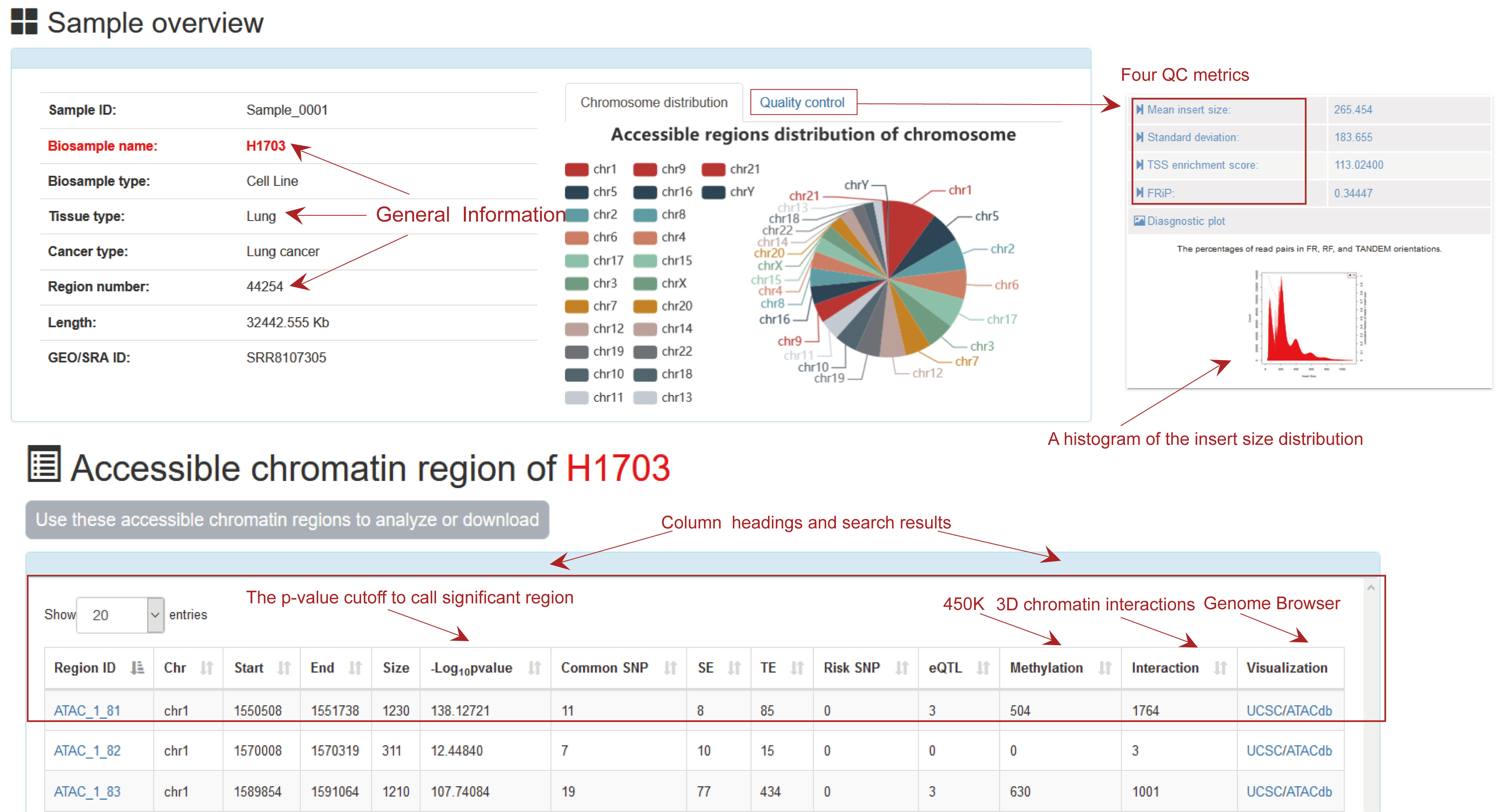
Search result of accessible chromatin regions for Sample_0001
Users determine the scope of the accessible chromatin regions query by determining the tissue type and Biosample Type for the results of interest. Brief information on the search results is displayed in a table on the result page.
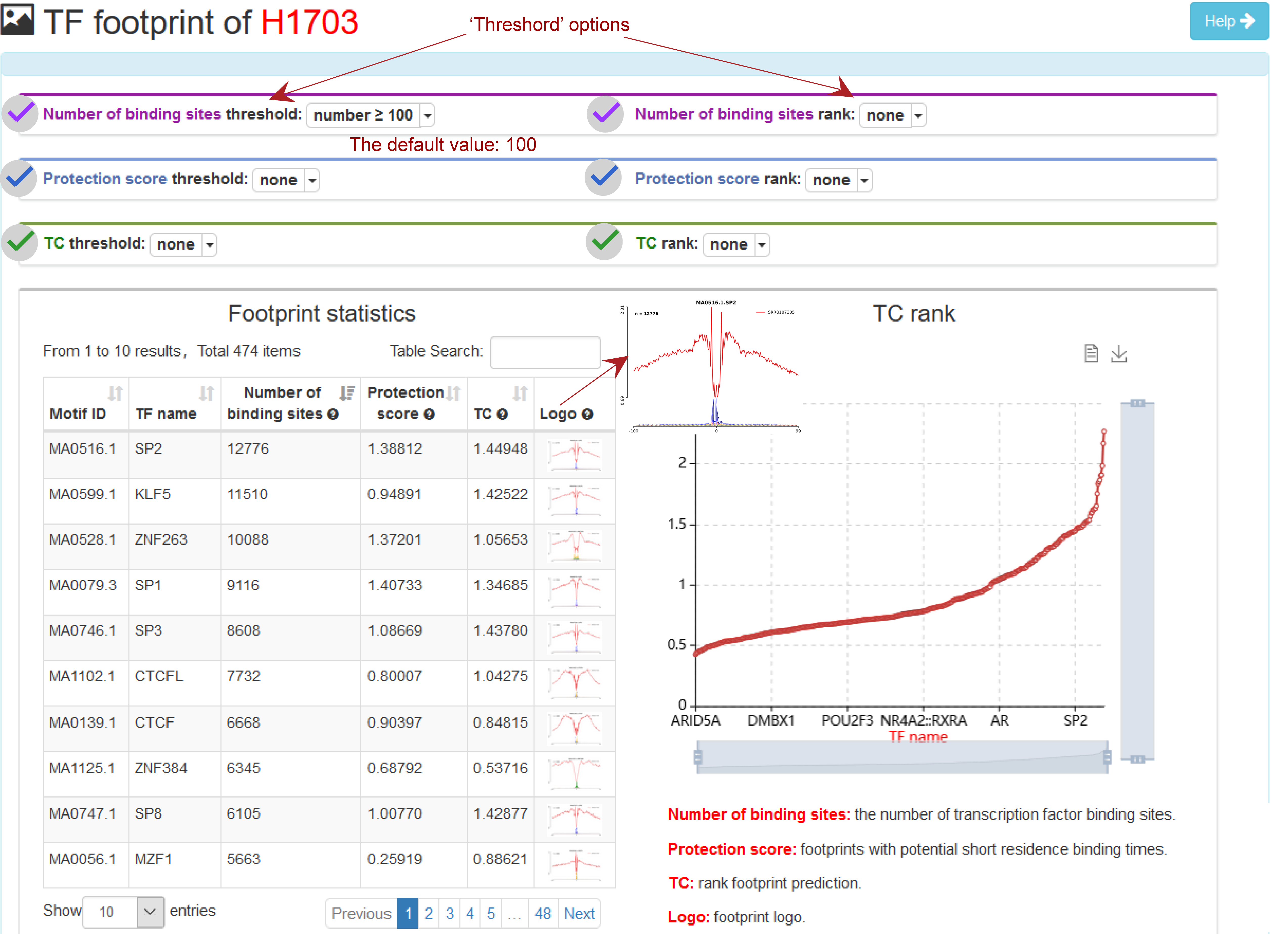
TF footprint search result for Sample_0001
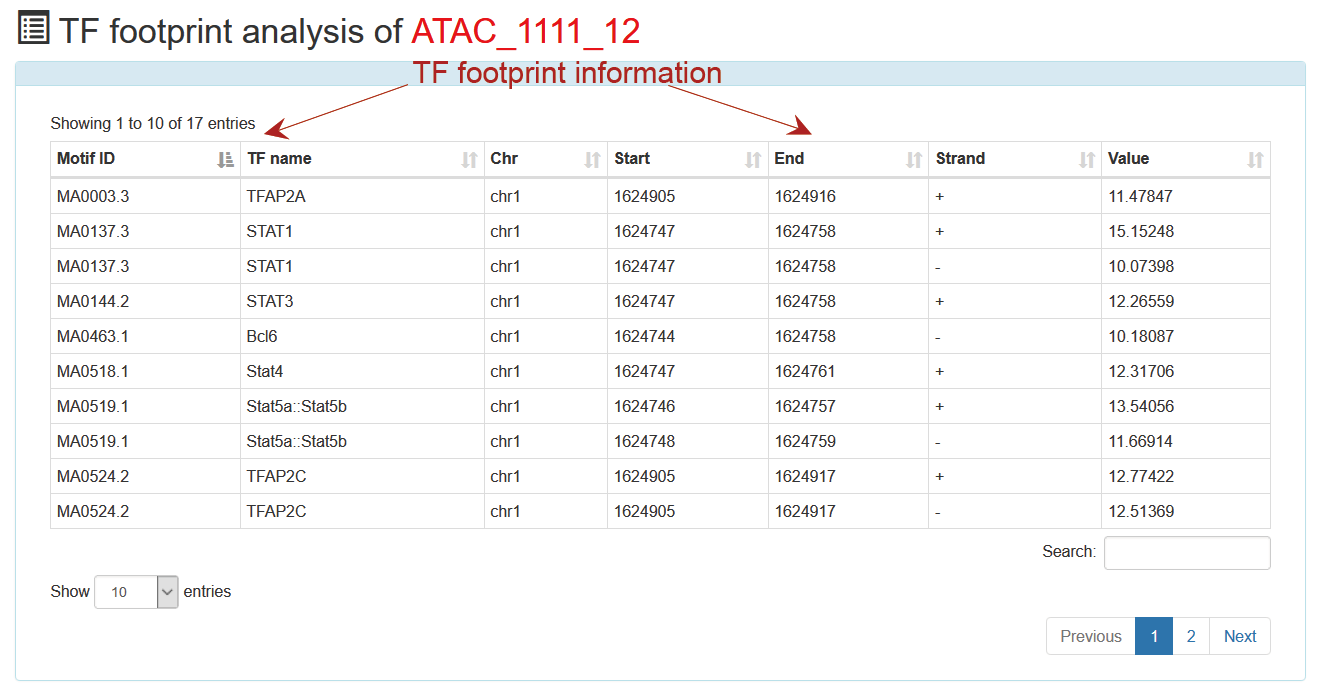
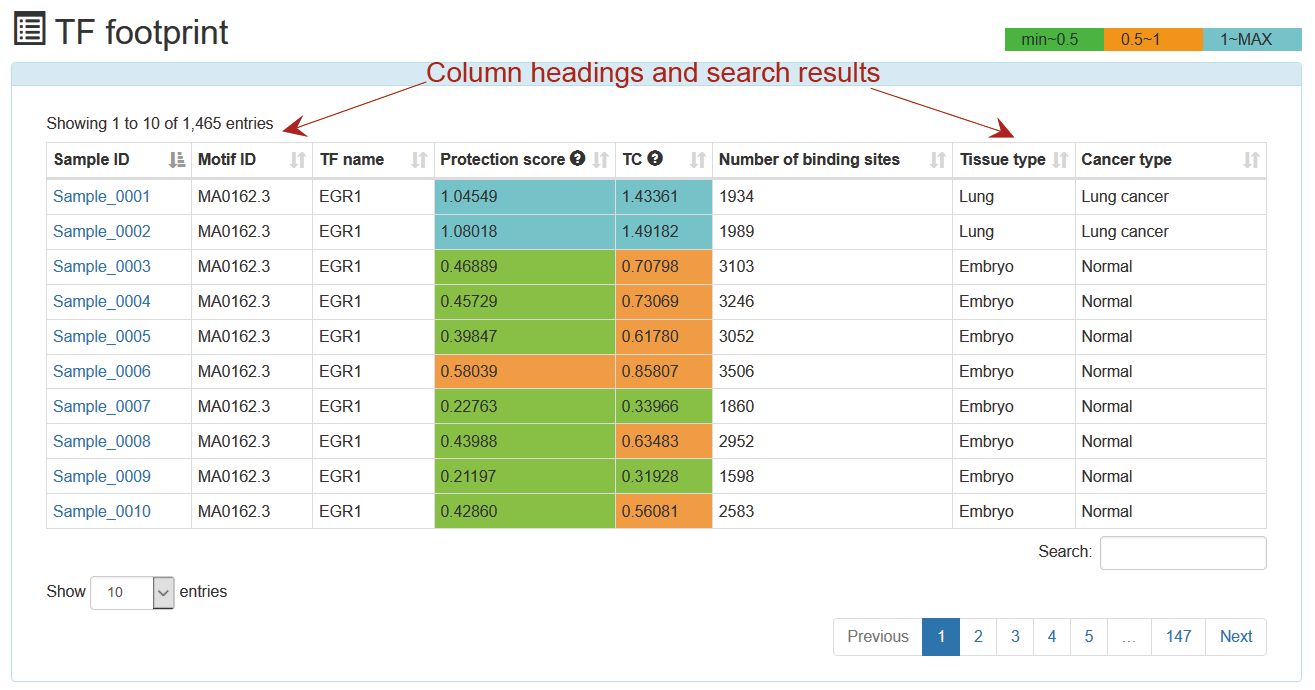
ATACdb predicts TFBS with footprints using HINT (41), which is based on hidden markov models. TFs with the Tag Count (TC), protection score, number of binding sites and footprint logo were identified for each sample. TC_score has been shown to be the best strategy for ranking footprint predictions. TF protection score can indicate footprints of TFs with potentially short binding times. The profiles for each motif, which can indicate the activity of TF intuitively. We have filtered out TFs with ≤ 10 binding sites. We have now added some new ‘Threshold’ options, including ‘Protection score threshold’, ‘TC threshold’ and ‘Number of binding sites threshold’, which allows users to set different thresholds to ensure TFs are high-activity and cell-type-specific. For example, we set a default threshold of the number of binding sites (the default value: 100).
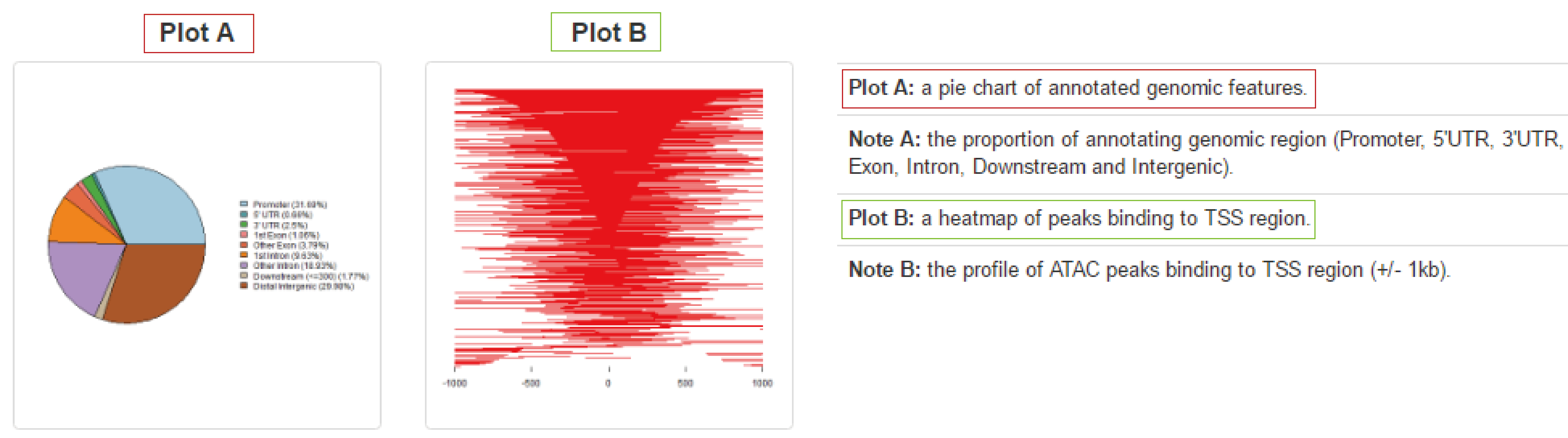
Peak annotation visualization
ATACdb implemented visualization function of peak annotation. We support visualization of ATAC-seq peaks in different ways, including displays of peaks coverage over chromosomes and profiles of peaks binding to TSS region.
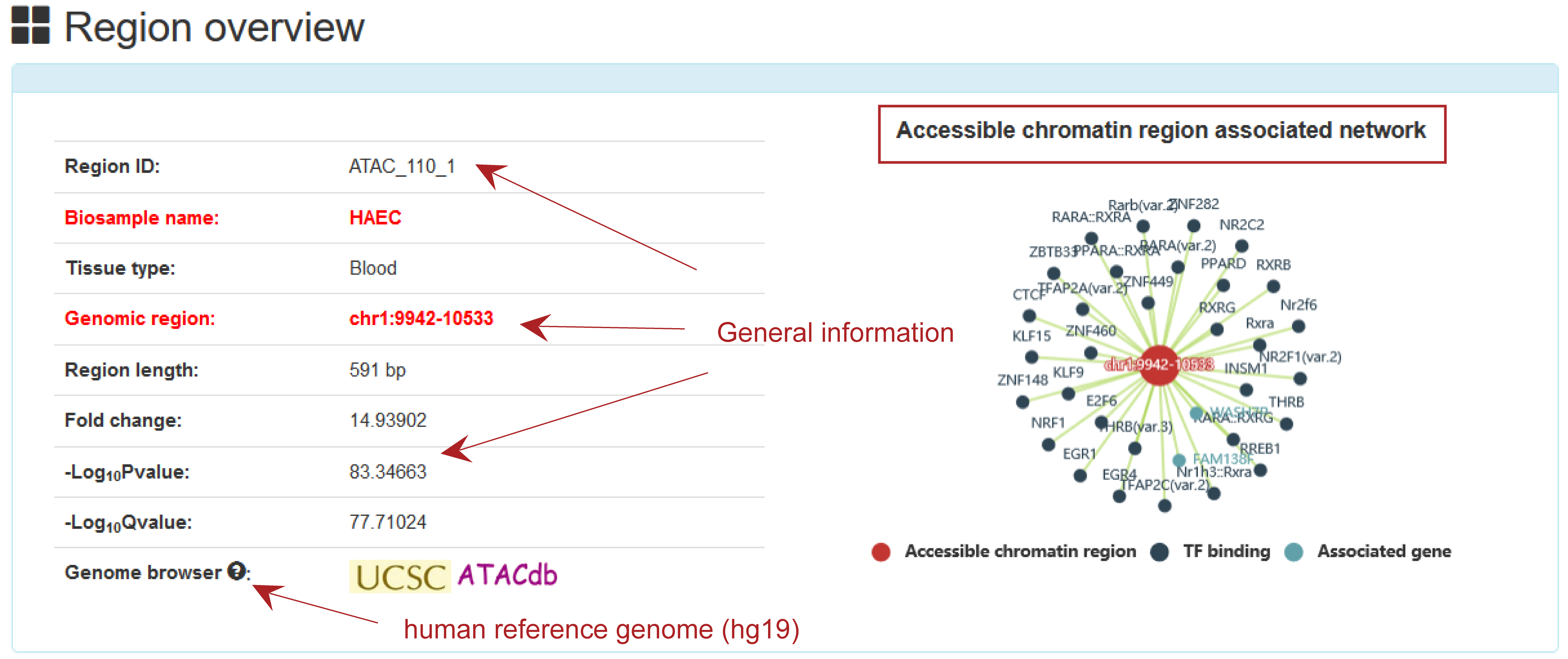
Users can click 'ATAC_1111_12'and the detail information about the region will be displayed on the next page.
Overview information includes Region ID, Biosample name, Tissue type, genomic region, region length, Fold change, -Log10Pvalue, -Log10Qvalue and Genome Browser.
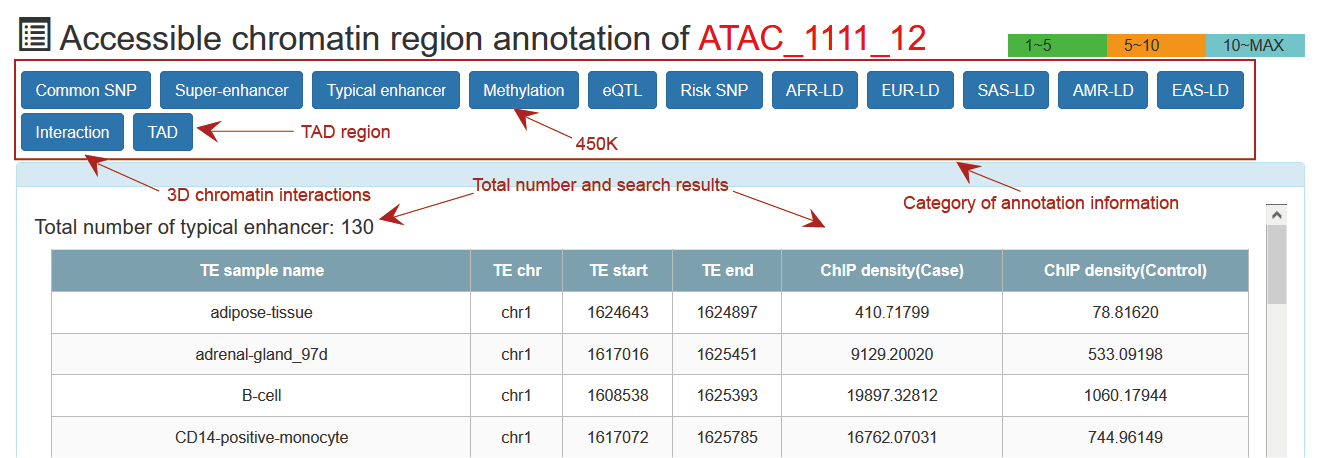
This is a detailed display of the 'ATAC_1111_12' annotation.
This is a detailed display of the 'ATAC_1111_12' TF footprint analysis.
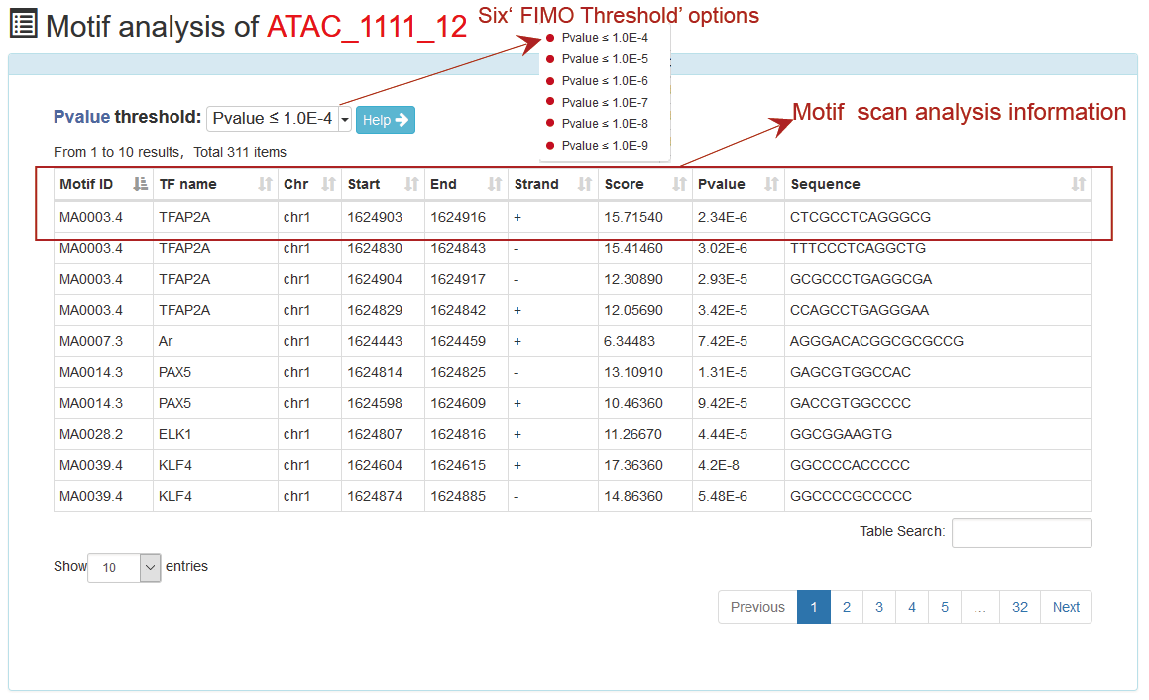
ATACdb identified individual candidate binding sites or protein motifs in a total of 52,078,883 accessible chromatin regions in ATACdb. We found that some motifs are short. They may not be found if users set a too stringent P-value of FIMO. Therefore, we identified DNA-binding sequence motifs with a P-value threshold of 1e−4, make sure that short motifs were also well represented in our database. We further added some ‘FIMO threshold’ options allowing users to select different parameters.
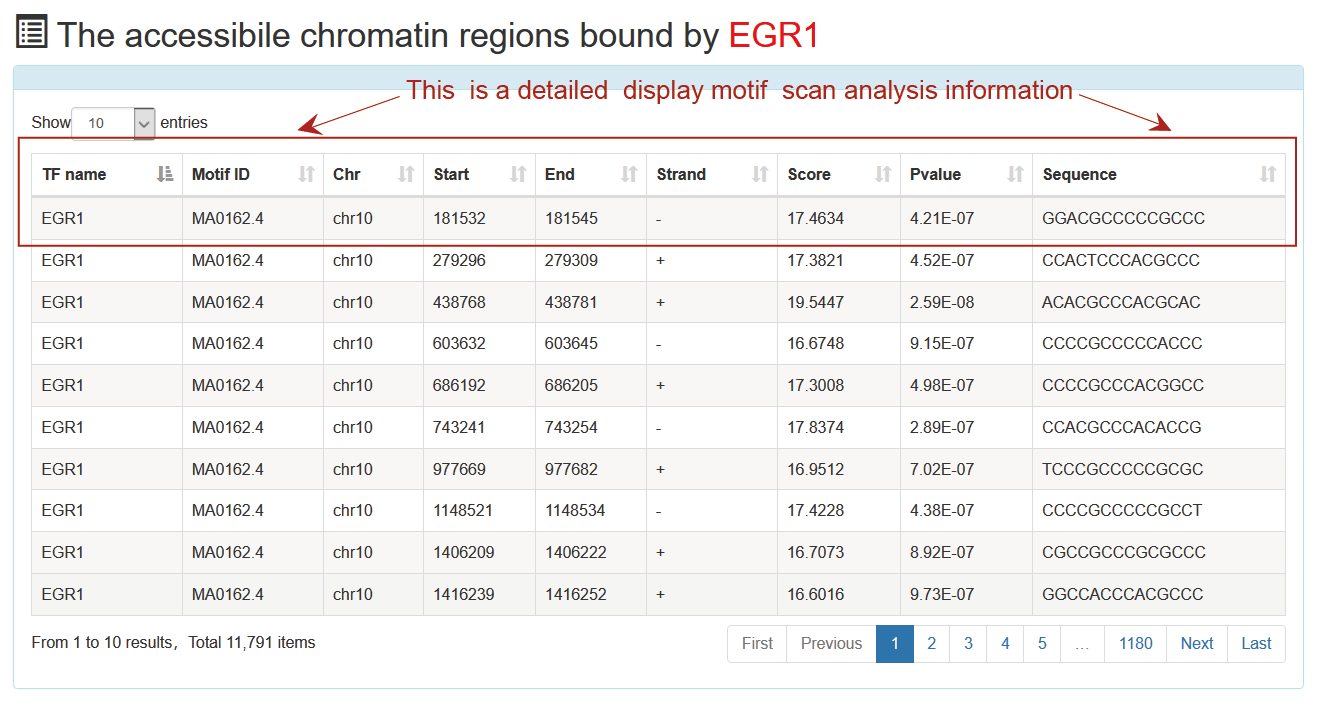
This is a detailed display of the 'ATAC_1111_12' motif scan analysis.
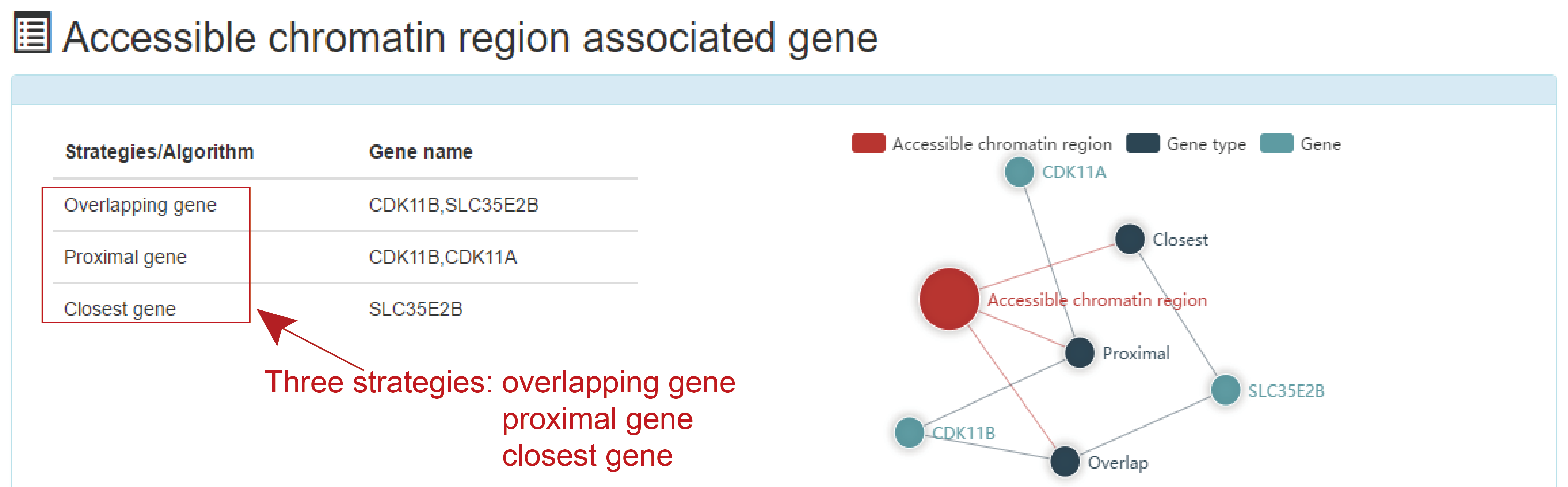
This is a detailed display of the 'ATAC_1111_12' associated genes.
Users submit a TF name and ATACdb will identify the accessible chromatin regions bound by the TF and their associated samples. TF related accessible chromatin regions are identified under two strategies:TF footprint and motif scan.
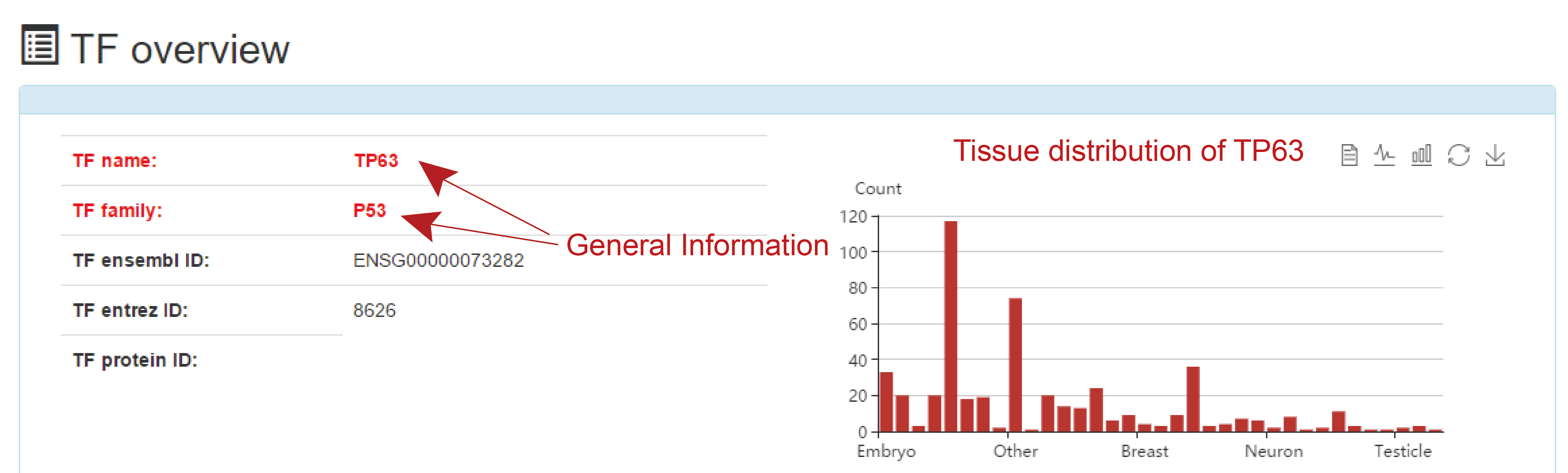
TF overview
This is a detailed display of the "TP63" footprint analysis result.
This is a detailed display of the "TP63" motif scan analysis information.
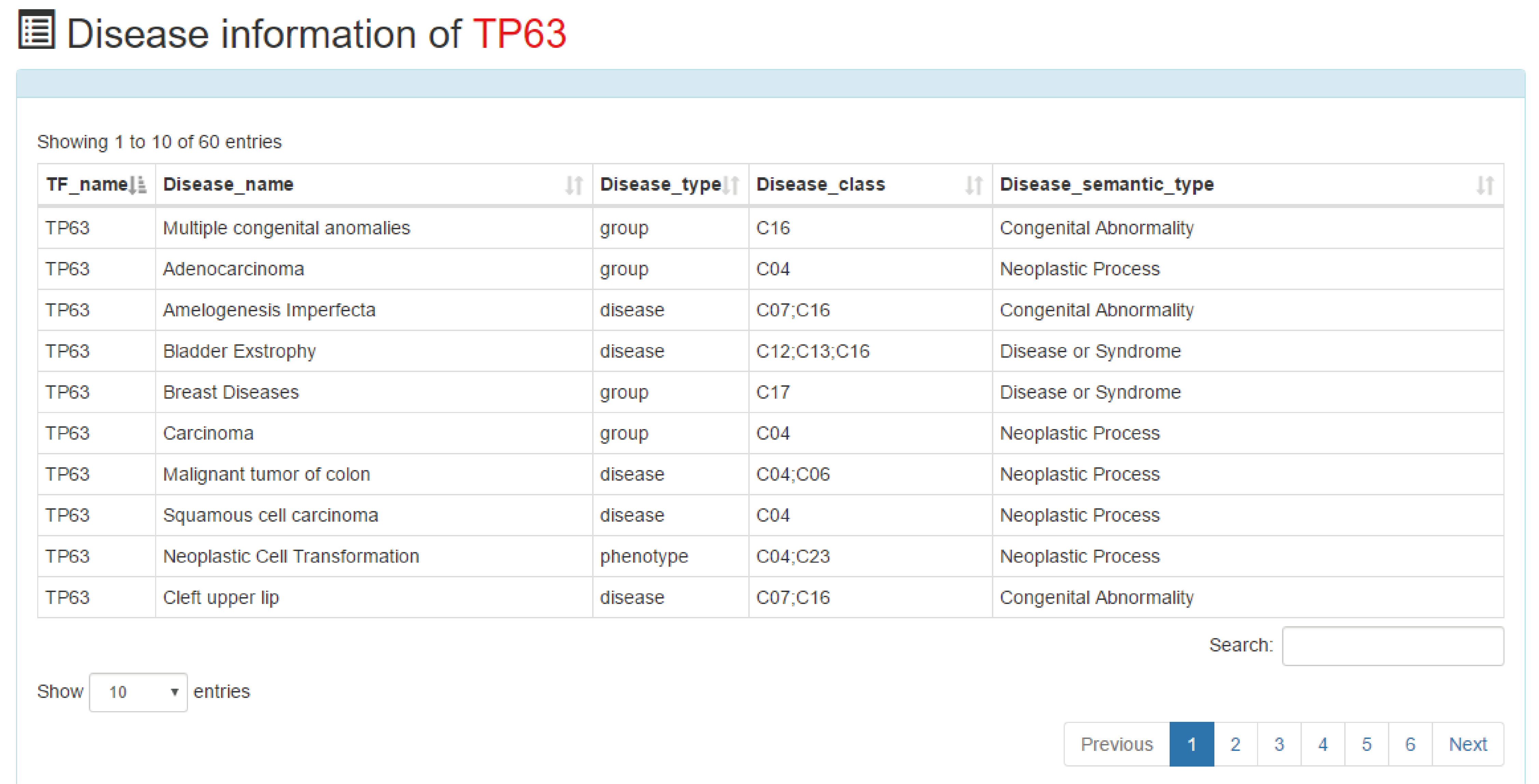
This is a detailed display of the disease information about "TP63".
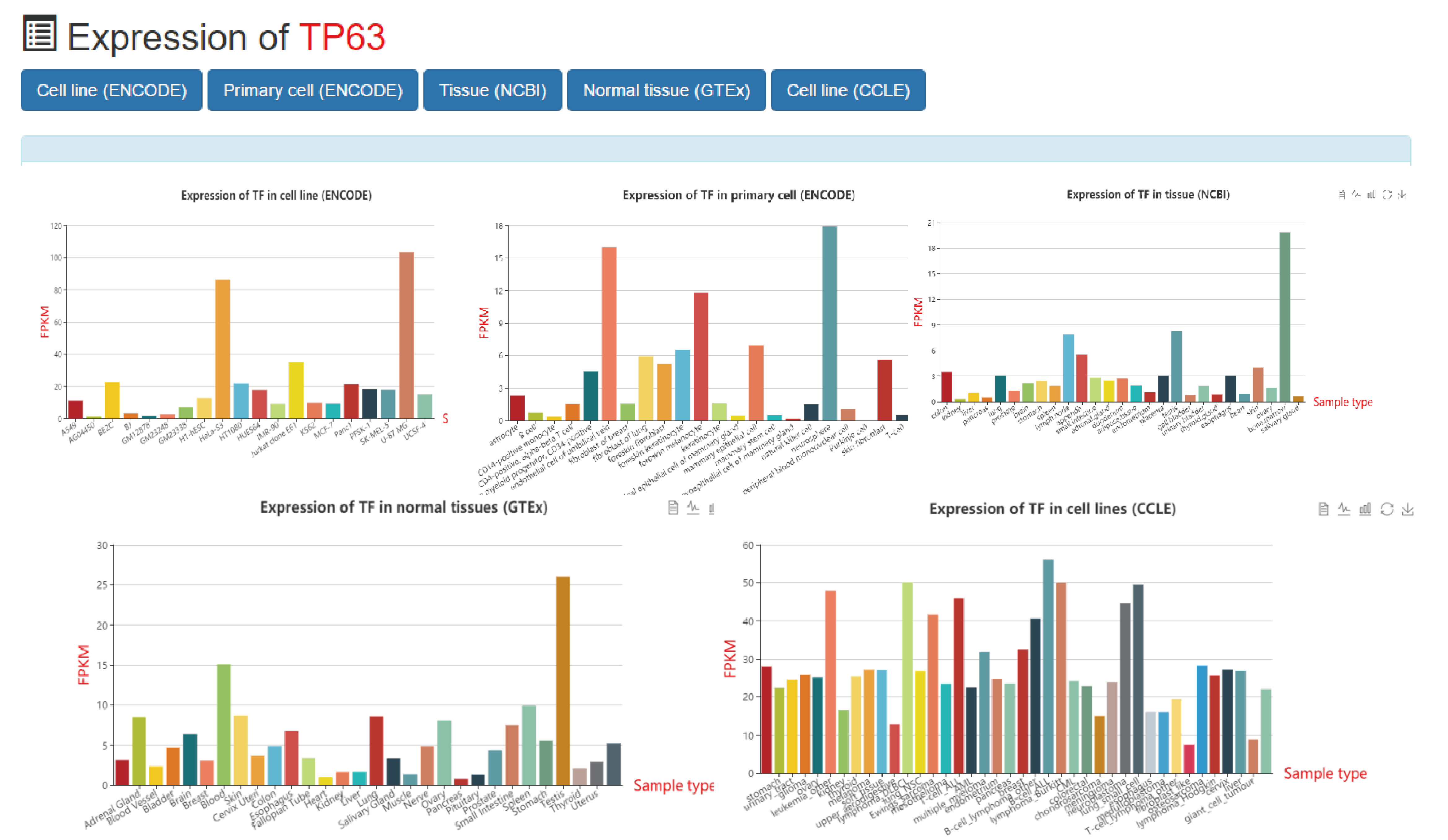
This is a detailed display of the "TP63" expression information.
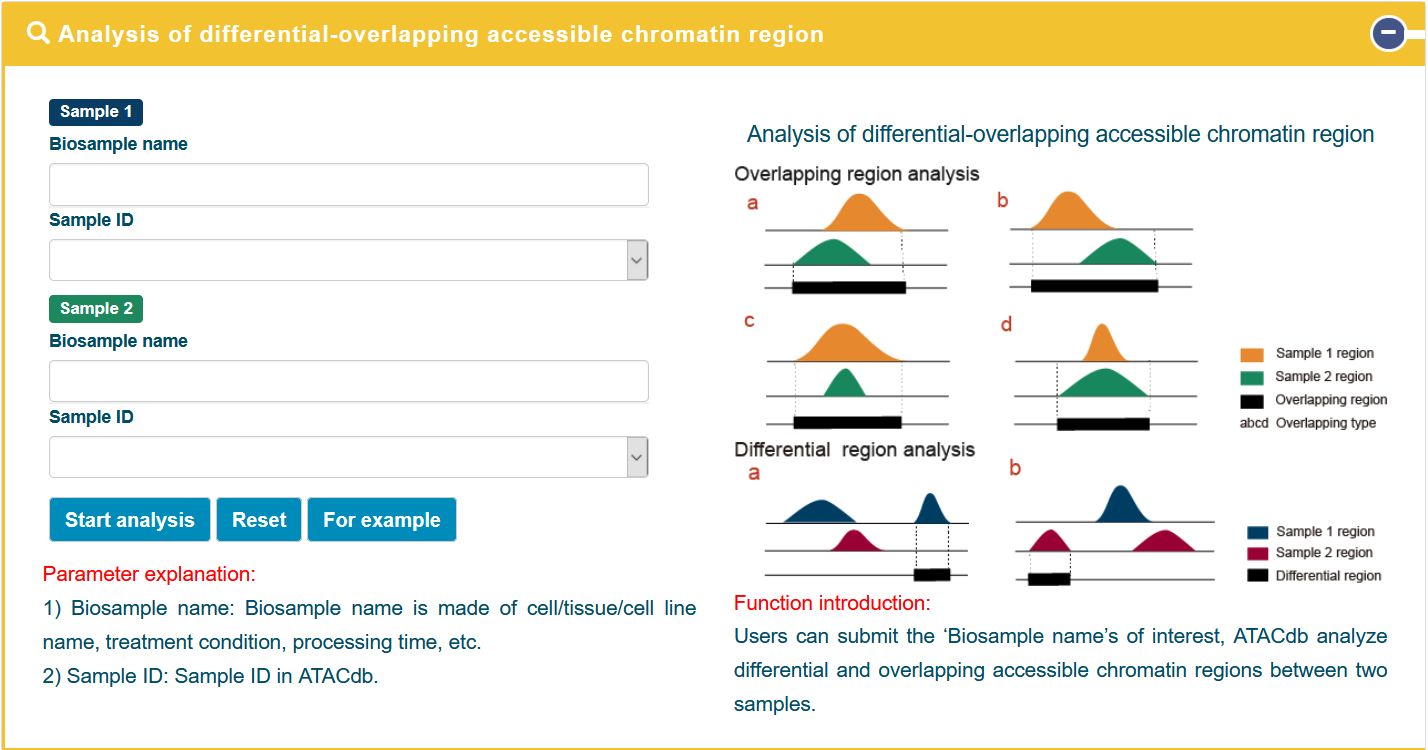
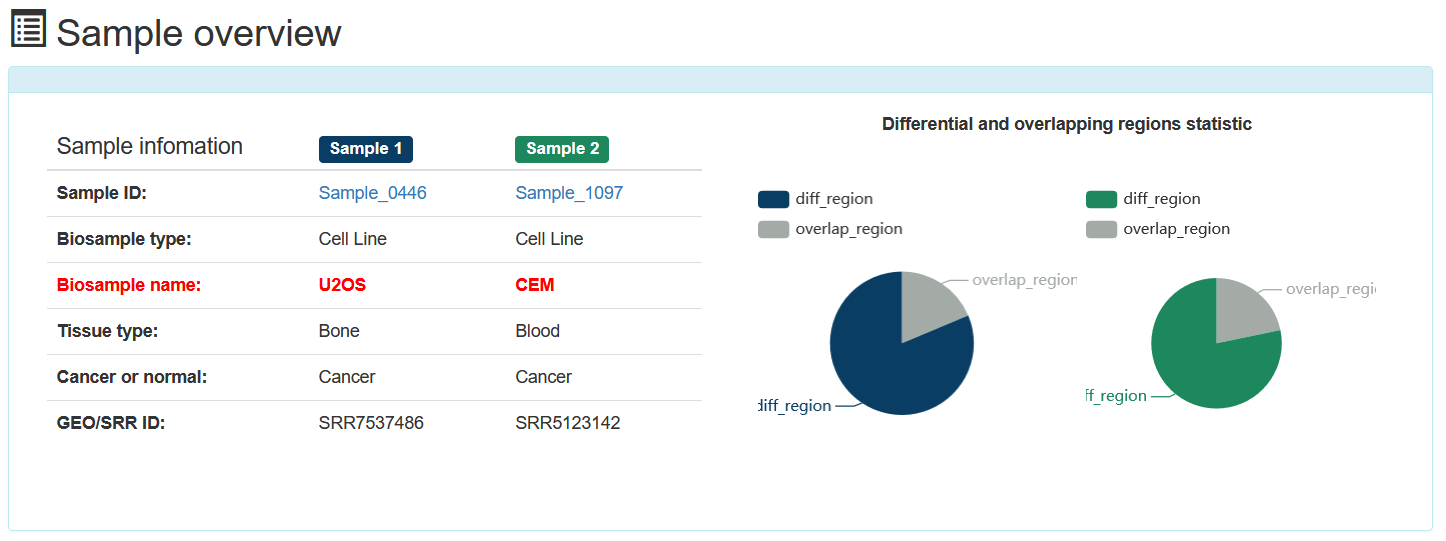
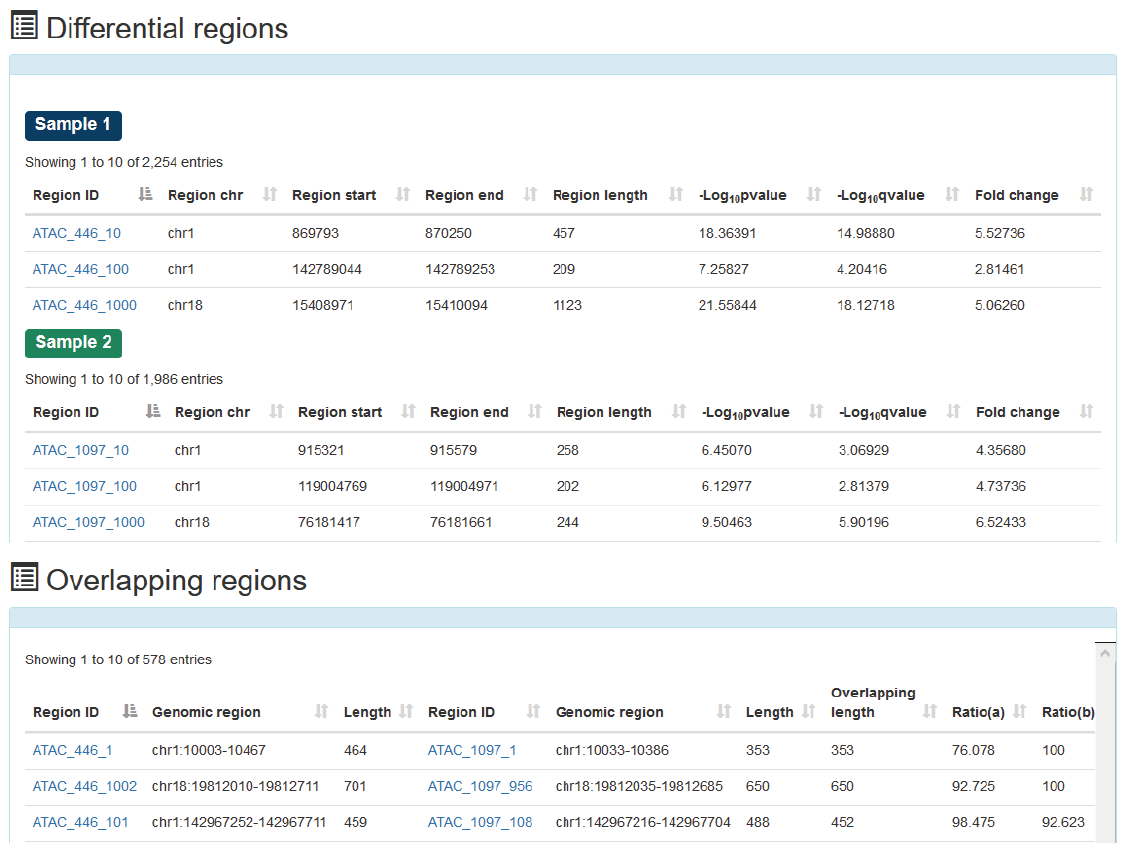
In the 'Differential-Overlapping-Region' analysis tool, users can submit the ‘Biosample name’s of interest, ATACdb analyze differential and overlapping accessible chromatin regions between two samples.
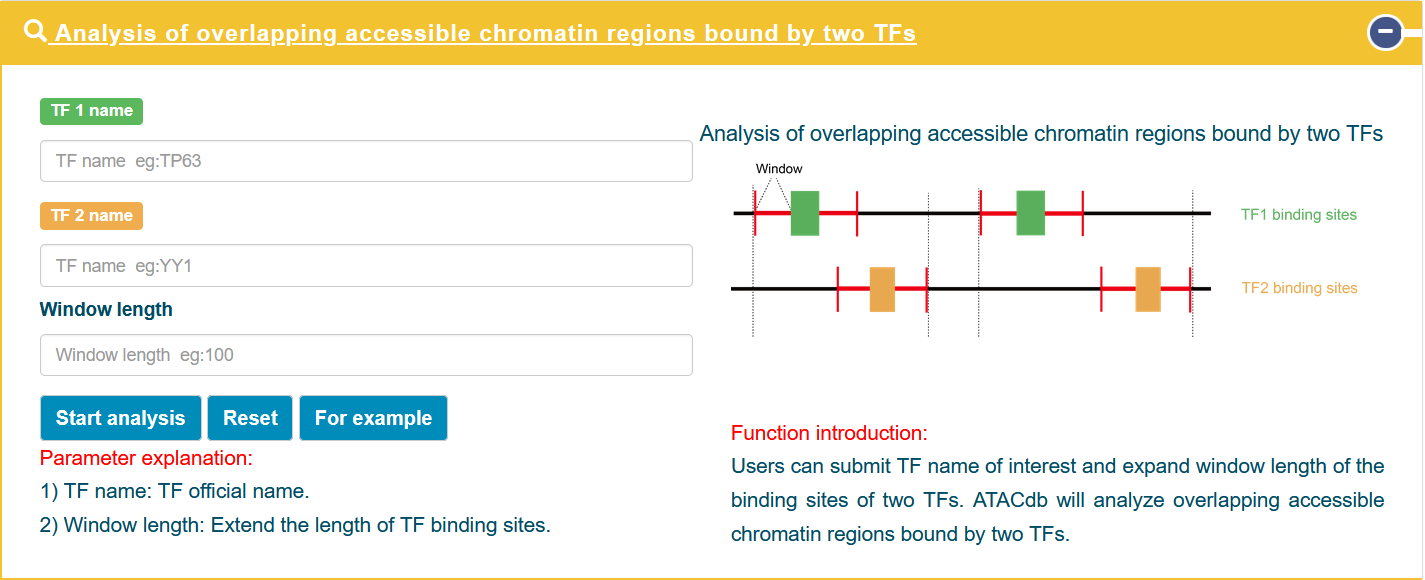
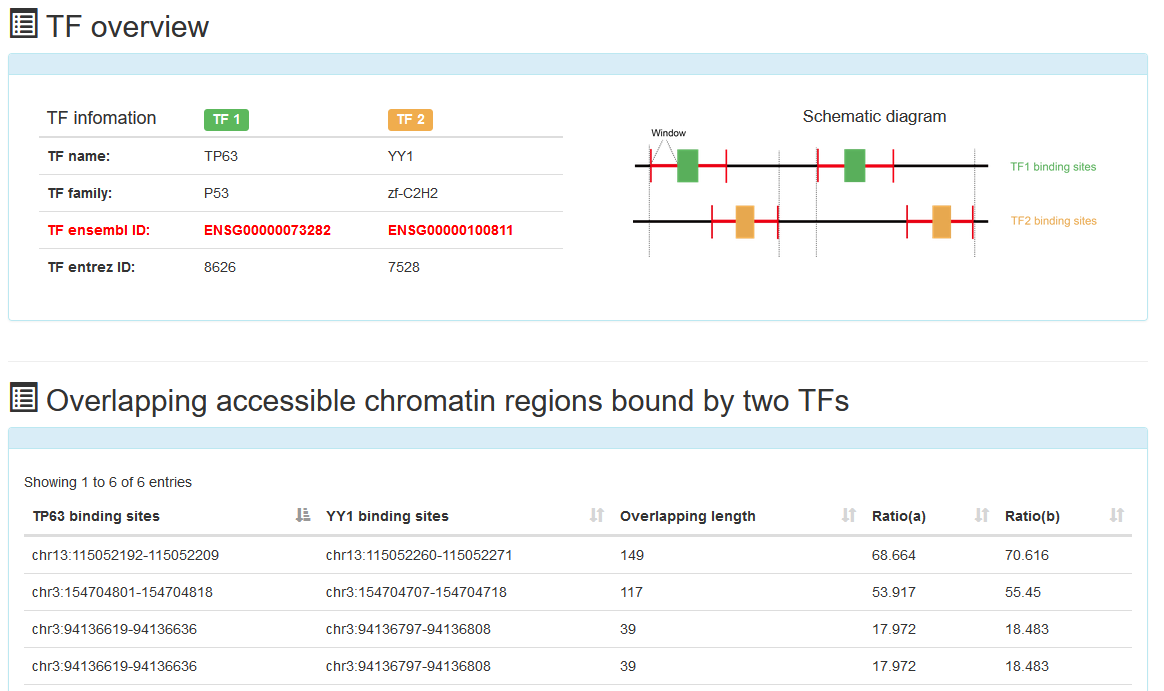
In the 'Overlapping accessible chromatin regions bound by two TFs' analysis tool, users can submit TF name of interest and expand window length of the binding sites of two TFs. ATACdb will analyze overlapping accessible chromatin regions bound by two TFs.
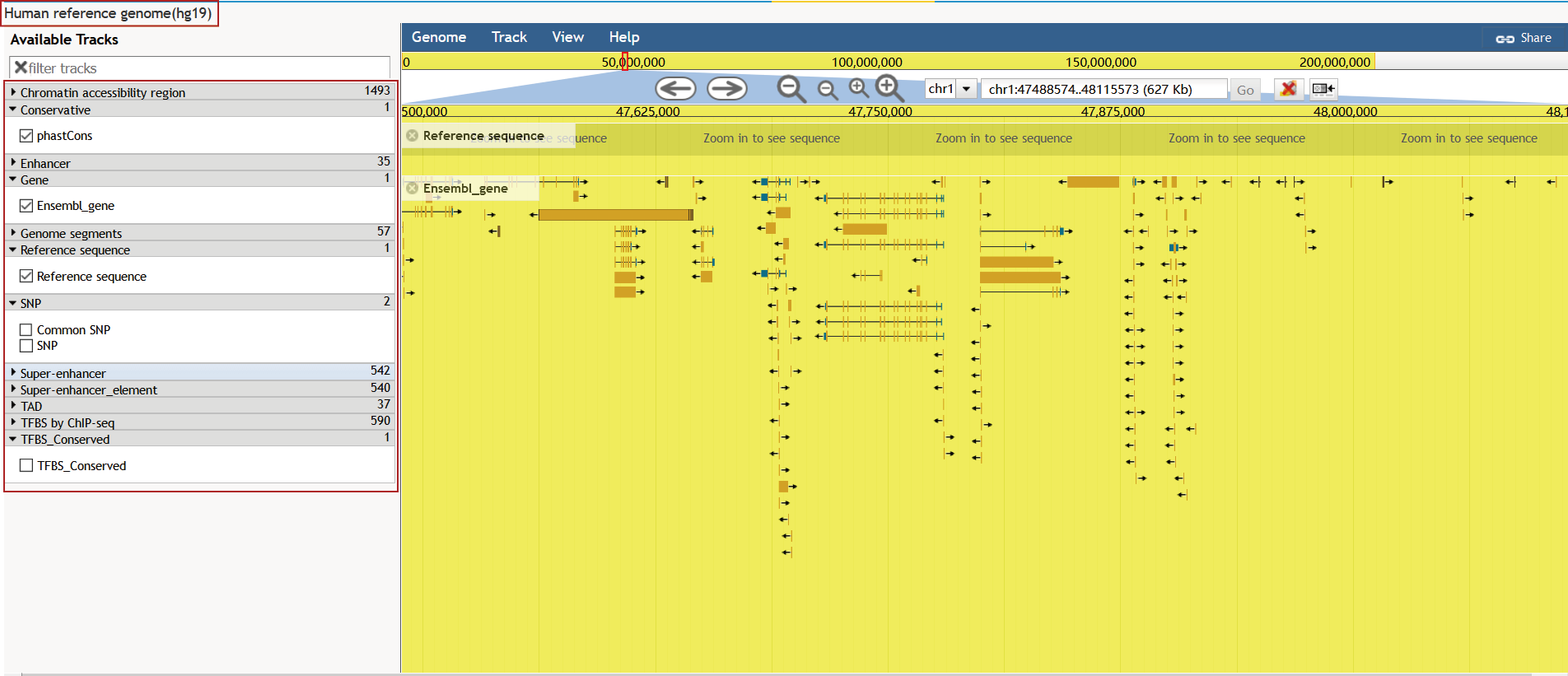
To help users view proximity information of accessible chromatin regions in genomes, wedeveloped a personalized genome browser using JBrowse with useful tracks. Users see the proximity of accessible chromatin regions to nearby genes, genome segments, SNPs, common SNPs, risk SNPs, enhancers, TFBS conserved, TFBS by ChIP-seq, and conservative score.
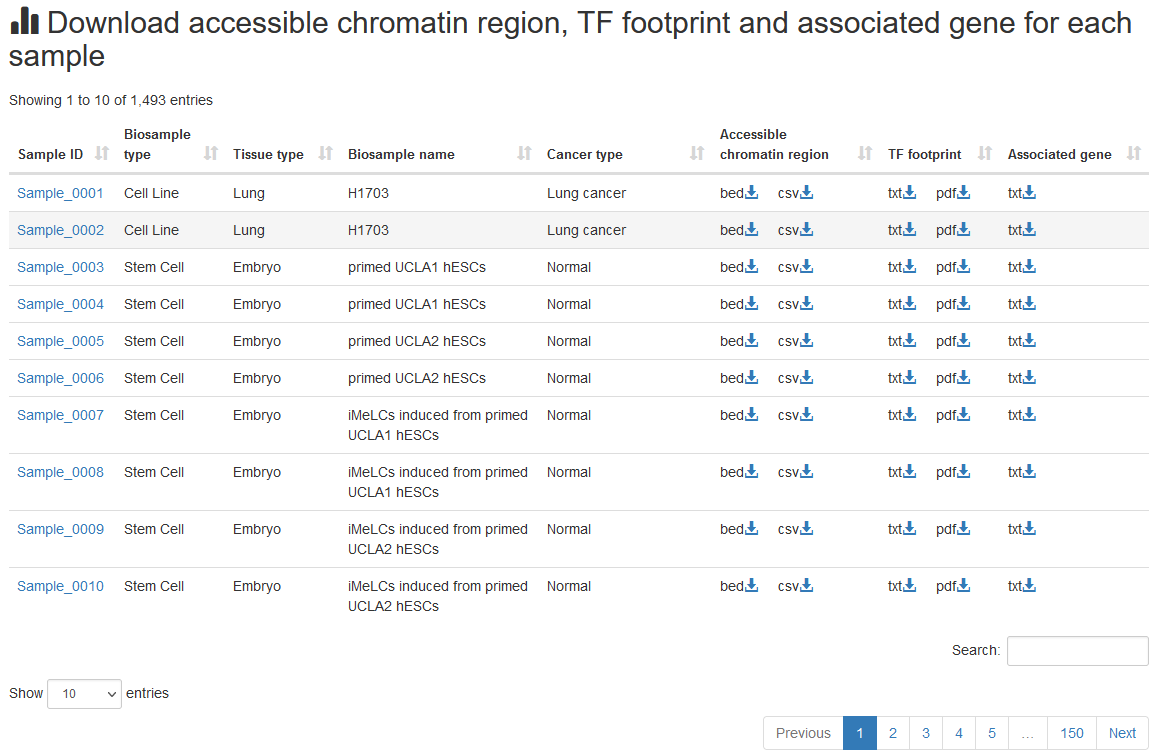
The data of accessible chromatin regions, TF footprints and motif scan results of all samples are provided for download in the ‘Download’ page. ATACdb supports the download of .bed format, .csv format, .pdf format and .txt format.
| ATACdb database contains 13 columns separated by tab: | Description |
|---|---|
| ATAC-seq | Assay for Transposase Accessible Chromatin with high-throughput sequencing. |
| TF footprint | A measure of TF occupancy, Tn5 footprint for motif sites falling into ATAC-seq data. TF footprint analysis using HINT, which parameters were set as described by Gusmao et al. |
| TC | TC (Tag Count) indicated the number of reads around putative transcription factor binding sites. We used TC to rank footprint prediction. |
| TF protection score | TF protection score was calculated by measuring the different Tn5 digestion numbers between flanking regions and TFBS, which indicated footprints with potential short residence binding times. |
| ‘Important’ in the context | To highlight the importance of this information, we added the label "important" on the detail page of accessible chromatin region. |
| Overlap rate | The overlap ratio between a certain sample of accessible chromatin regions and another sample of accessible chromatin regions. |
| Biosample type | Cell type classification of samples. |
| Tissue type | Samples tissue type. |
| Cancer type | Samples cancer type. |
| Biosample name | Biosample name is made of cell/tissue/cell line name, treatment condition, processing time, etc. |
| Chromosome | Chromosome. |
| Start position | The accessible chromatin region is at the start of the chromosome. |
| End position | The accessible chromatin region is at the end of the chromosome. |
Reply: Because there is no corresponding functional annotation or no significant annotation results for this TF/gene.
Reply: We provided four different QC metrics of ATAC-seq samples, including mean insert size and corresponding standard deviation of paired-end libraries using Picard, TSS enrichment score and FRiP using the ENCODE consortium. We preferred the mean insert size as a superior metric of quality assessment, because it was estimated after trimming off the outliers in from the original insert-size distribution. The TSS enrichment score indicated the average depth of the TSS of genes and the FRiP indicated fraction of mapped reads falling into the peak regions.
Reply: ATACdb has advanced storage technology and sufficient bandwidth to meet the needs of most users for the speed of web page loading. However, it is not excluded that few users have poor user experience due to network reasons.
Reply: Motif scan can be used to scan sequences of interest to predict TF binding sites. TF footprints analysis reveals the presence of the DNA binding protein at each site, analogous to DNase digestion footprints. Compared to motif scan, footprint identification is an accurate way to identify TFs associcated with the accessible chromatin regions.
The ATACdb website runs on a Linux-based Apache Web server 2.4.6 (http://www.apache.org). The database was developed using MySQL 5.7.27 (http://www.mysql.com). PHP 5.6.40 (http://www.php.net) was used for server-side scripting. The ATACdb web interface was built using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery.com). ECharts (http://echarts.baidu.com) was used to be a graphical visualization framework. This database has been tested using Mozilla Firefox, Google Chrome and Internet Explorer web browsers.
ATACdb is freely available to the research community at (http://www.licpathway.net/ATACdb) and quires no registration or login.