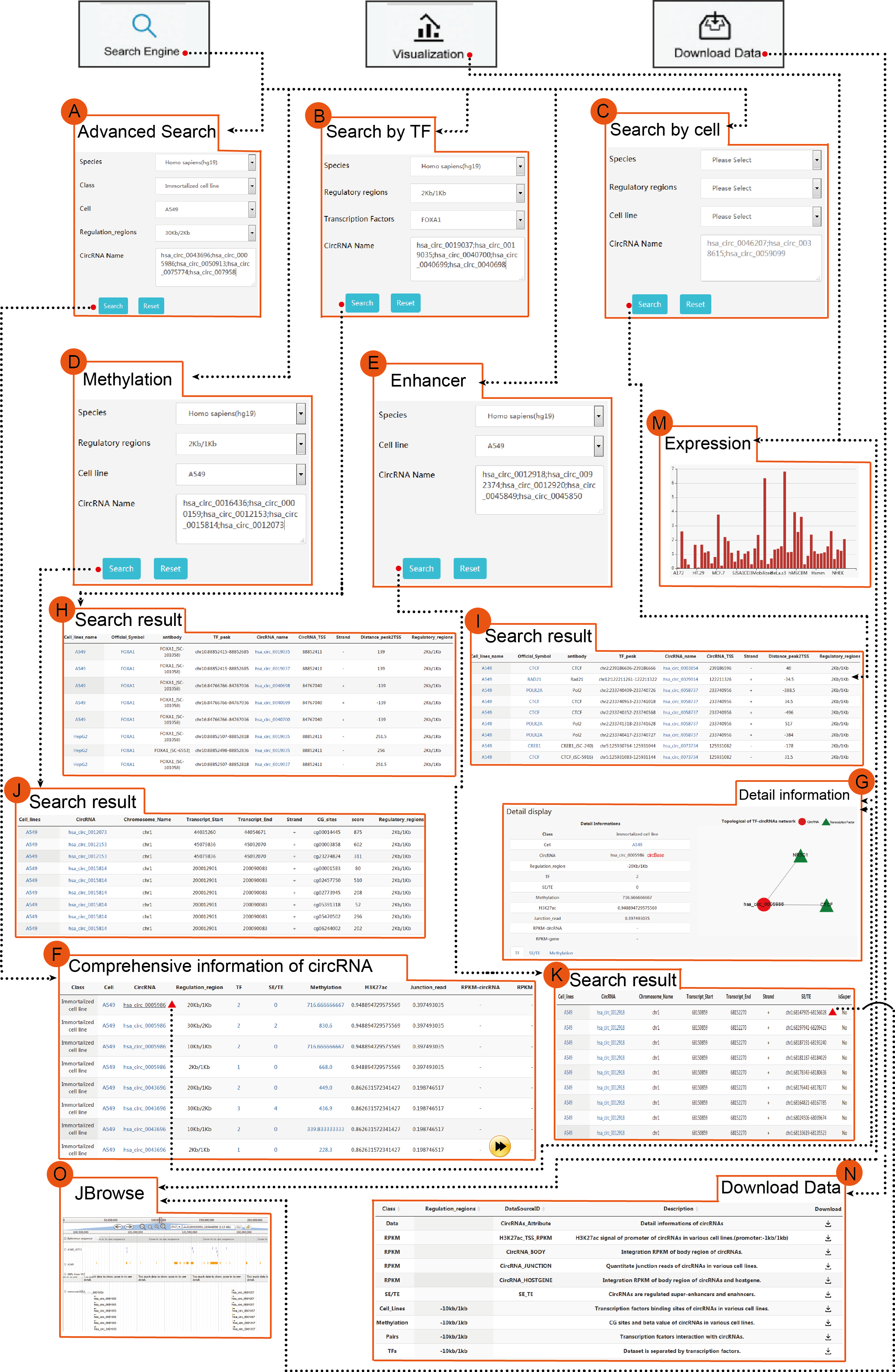
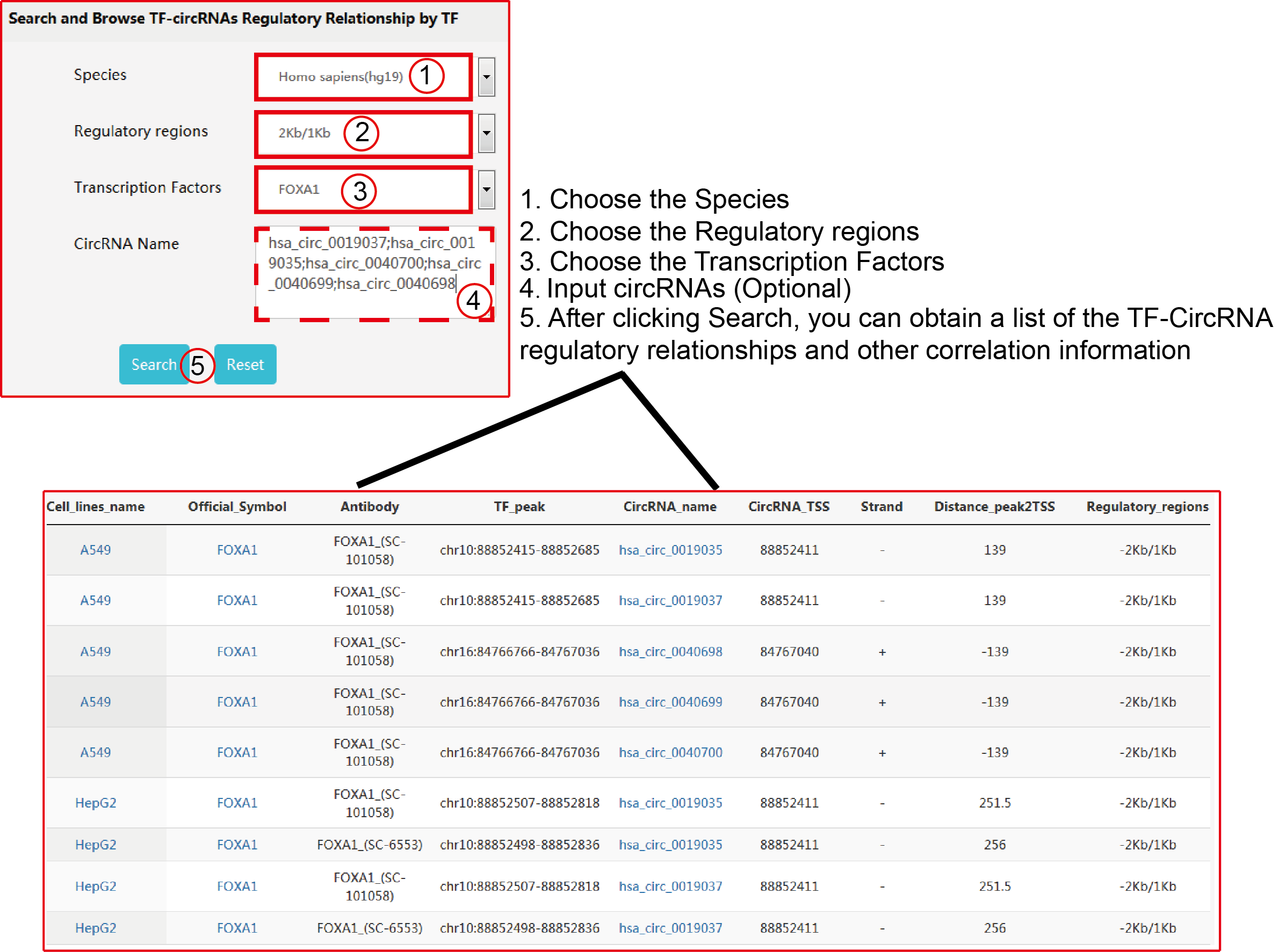
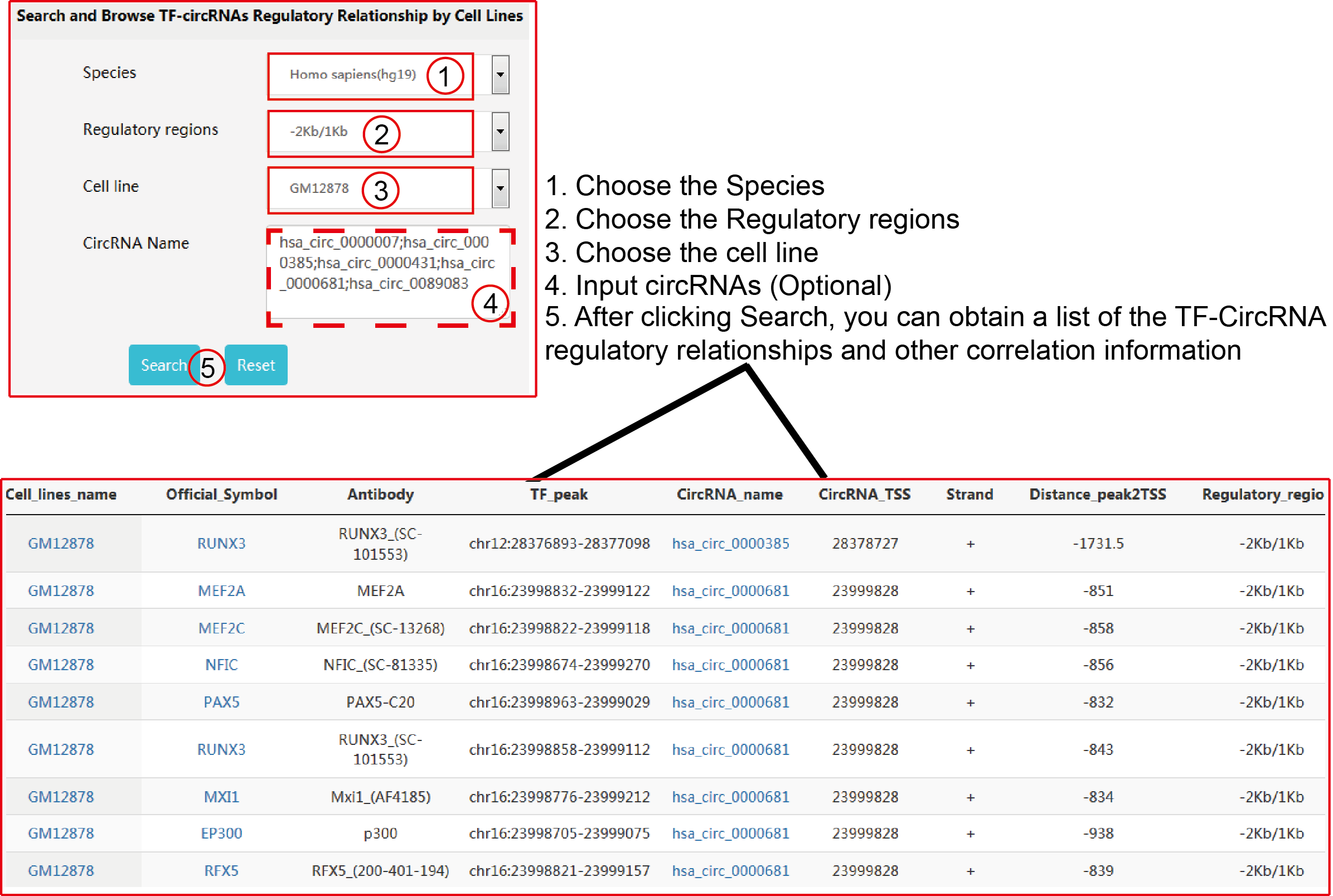
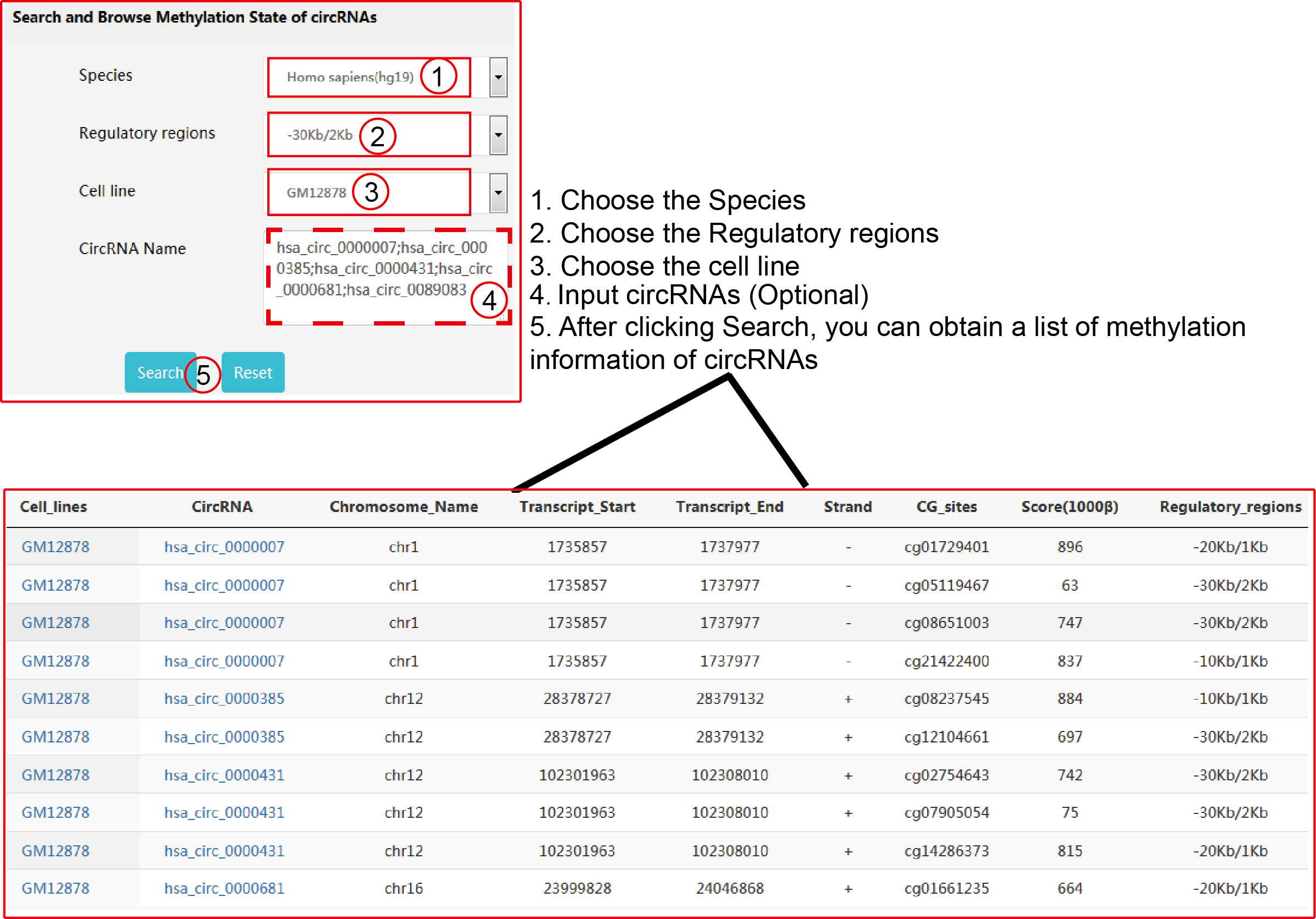
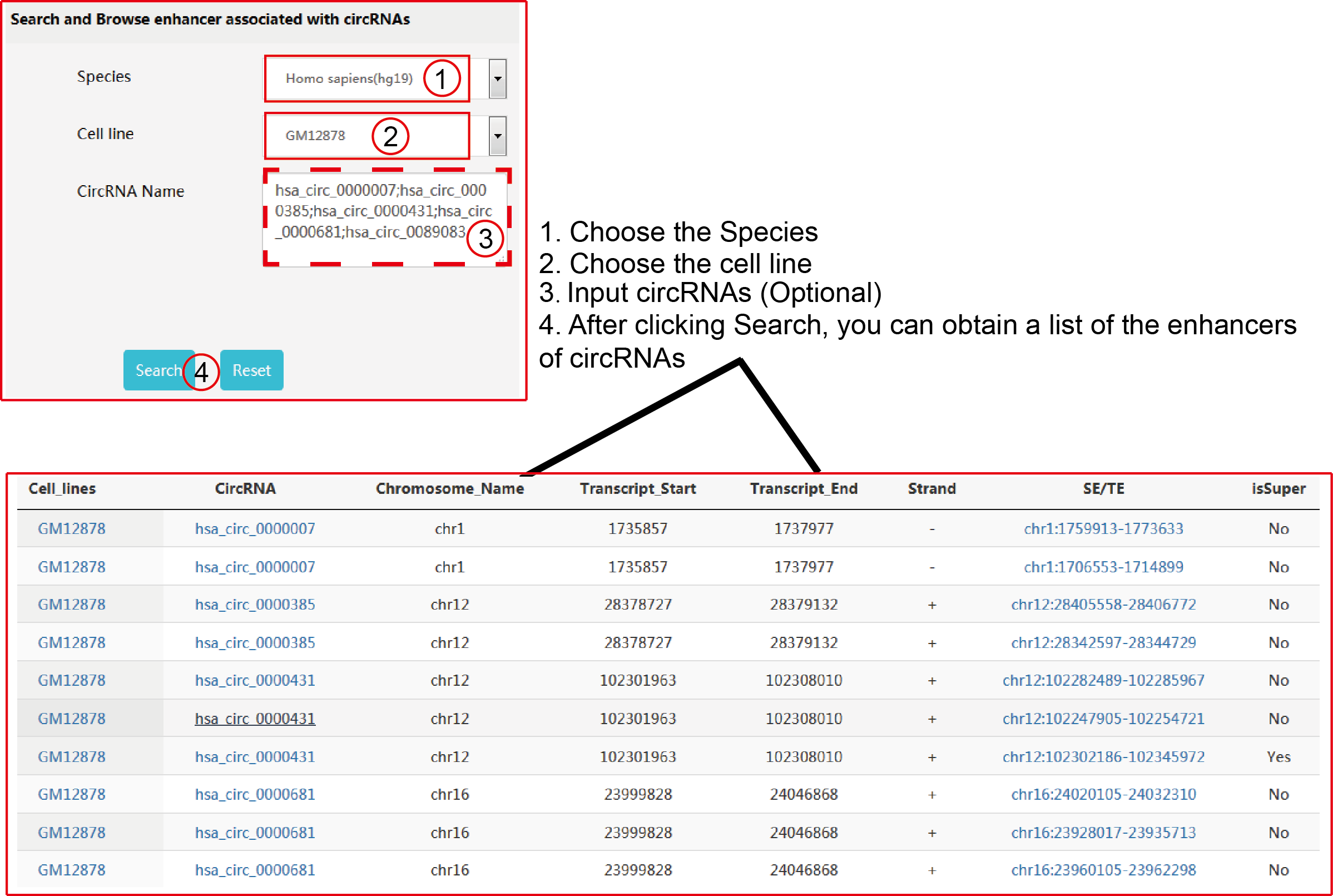
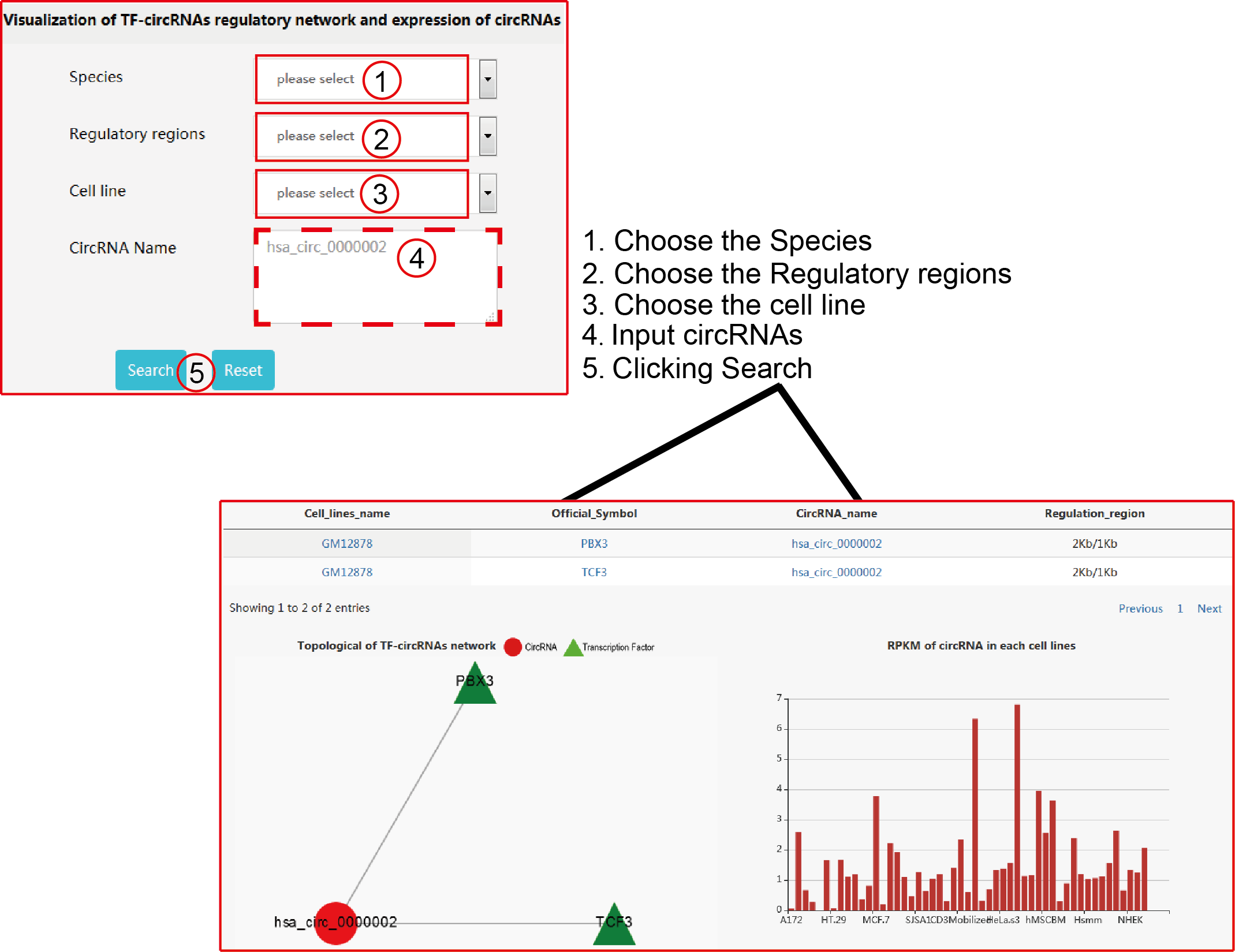
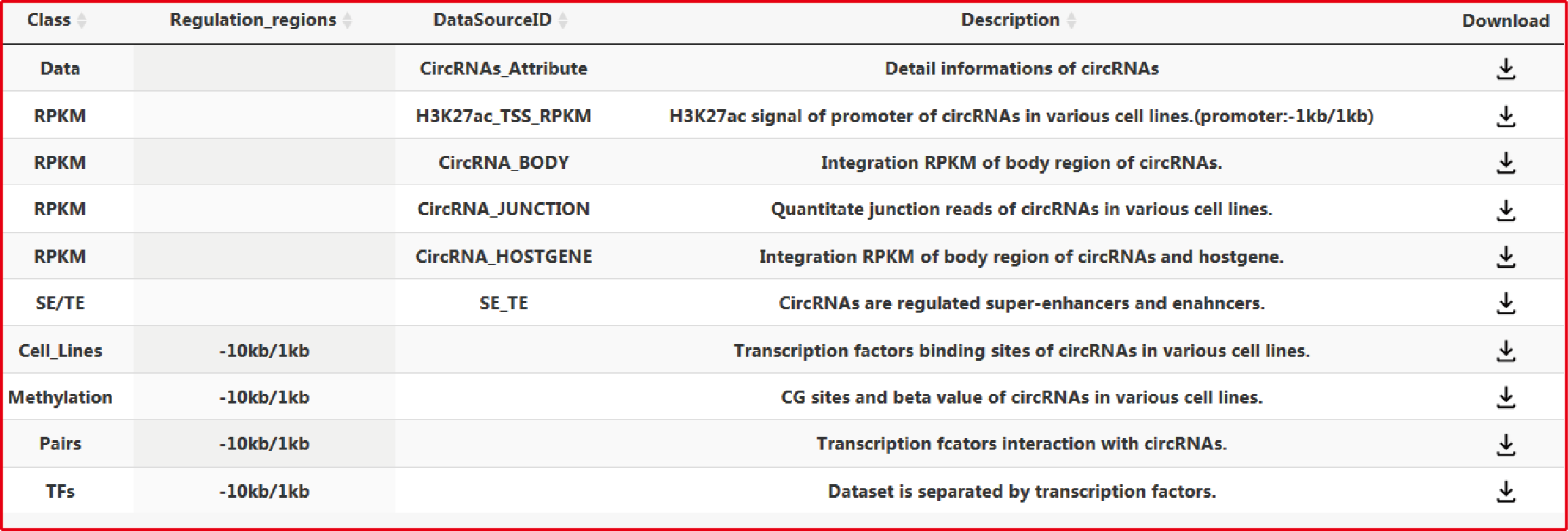
TRCirc provides a comprehensive data source for integrating current transcription factors binding sites and circRNA annotations. The current release of TRCirc contains approximately 92375 human circRNAs and 690 ChIP-seq data sets across 91 human cell lines for 161 unique regulatory factors.Moreover, TRCirc provides other information of circRNAs, such as enhancer, methylation level, H3K27ac signal and expression. TF2LncRNA is an efficient and easy-to-use web-based tool.